Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch
Xiao, H., Edwards, T.E., Ferre-D'Amare, A.R.(2008) Chem Biol 15: 1125-1137
- PubMed: 18940672
- DOI: https://doi.org/10.1016/j.chembiol.2008.09.004
- Primary Citation of Related Structures:
3EGZ - PubMed Abstract:
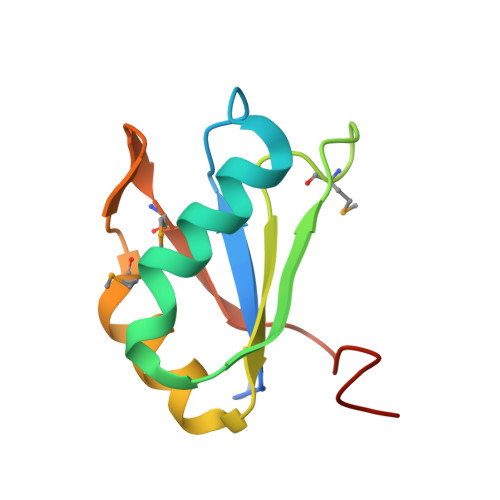
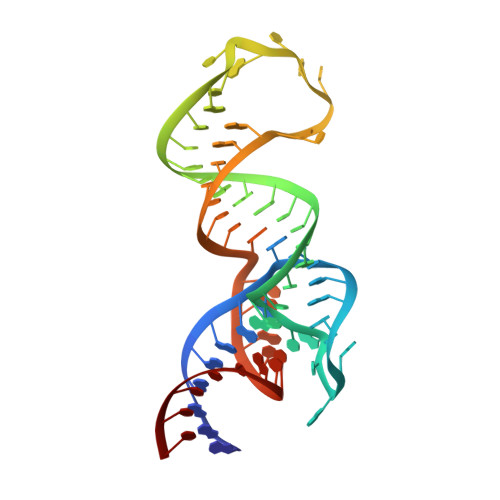
The tetracycline aptamer is an in vitro selected RNA that binds to the antibiotic with the highest known affinity of an artificial RNA for a small molecule (Kd approximately 0.8 nM). It is one of few aptamers known to be capable of modulating gene expression in vivo. The 2.2 A resolution cocrystal structure of the aptamer reveals a pseudoknot-like fold formed by tertiary interactions between an 11 nucleotide loop and the minor groove of an irregular helix. Tetracycline binds within this interface as a magnesium ion chelate. The structure, together with previous biochemical and biophysical data, indicates that the aptamer undergoes localized folding concomitant with tetracycline binding. The three-helix junction, h-shaped architecture of this artificial RNA is more complex than those of most aptamers and is reminiscent of the structures of some natural riboswitches.
- Howard Hughes Medical Institute, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, WA 98109-1024, USA.
Organizational Affiliation: