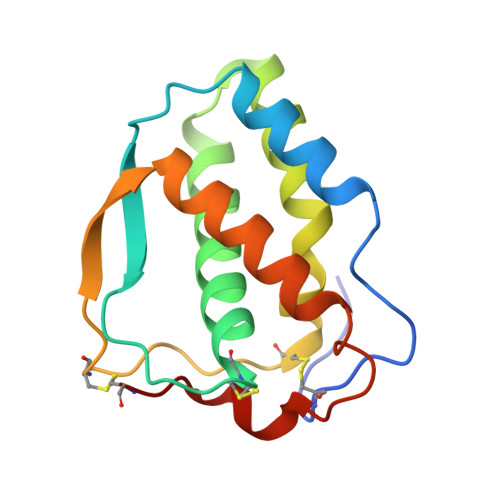
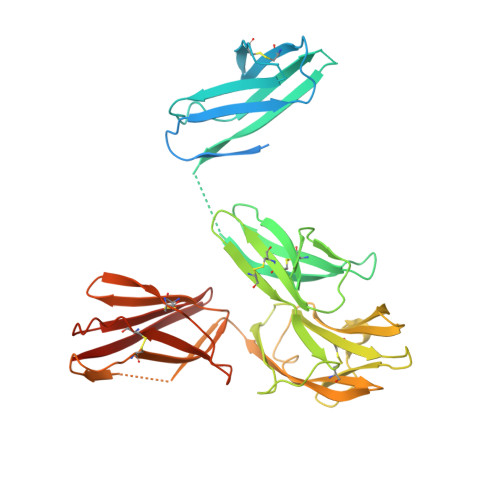
Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex.
Verstraete, K., Vandriessche, G., Januar, M., Elegheert, J., Shkumatov, A.V., Desfosses, A., Van Craenenbroeck, K., Svergun, D.I., Gutsche, I., Vergauwen, B., Savvides, S.N.(2011) Blood 118: 60-68
- PubMed: 21389326
- DOI: https://doi.org/10.1182/blood-2011-01-329532
- Primary Citation of Related Structures:
3QS7, 3QS9 - PubMed Abstract:
The class III receptor tyrosine kinase (RTKIII) Fms-like tyrosine kinase receptor 3 (Flt3) and its cytokine ligand (FL) play central roles in hematopoiesis and the immune system, by establishing signaling cascades crucial for the development and homeostasis of hematopoietic progenitors and antigen-presenting dendritic cells. However, Flt3 is also one of the most frequently mutated receptors in hematologic malignancies and is currently a major prognostic factor and clinical target for acute myeloid leukemia. Here, we report the structural basis for the Flt3 ligand-receptor complex and unveil an unanticipated extracellular assembly unlike any other RTKIII/V complex characterized to date. FL induces dimerization of Flt3 via a remarkably compact binding epitope localized at the tip of extracellular domain 3 of Flt3, and it invokes a ternary complex devoid of homotypic receptor interactions. Comparisons of Flt3 with homologous receptors and available mutagenesis data for FL have allowed us to rationalize the unique features of the Flt3 extracellular assembly. Furthermore, thermodynamic dissection of complex formation points to a pronounced enthalpically driven binding event coupled to an entropic penalty. Together, our data suggest that the high-affinity Flt3:FL complex is driven in part by a single preformed binding epitope on FL reminiscent of a "lock-and-key" binding mode, thereby setting the stage for antagonist design.
- Unit for Structural Biology, Laboratory for Protein Biochemistry and Biomolecular Engineering (L-ProBE), Ghent University, Ghent, Belgium.
Organizational Affiliation: