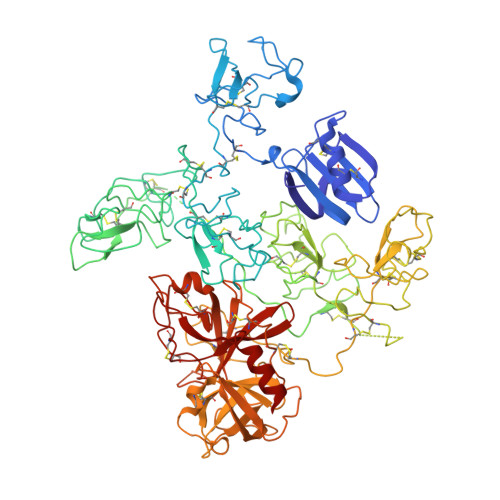
The X-ray crystal structure of full-length human plasminogen
Law, R.H.P., Caradoc-Davies, T., Cowieson, N., Horvath, A.J., Quek, A.J., Encarnacao, J.A., Steer, D., Cowan, A., Zhang, Q., Lu, B.G.C., Pike, R.N., Smith, A.I., Coughlin, P.B., Whisstock, J.C.(2012) Cell Rep 1: 185-190
- PubMed: 22832192
- DOI: https://doi.org/10.1016/j.celrep.2012.02.012
- Primary Citation of Related Structures:
4DUR, 4DUU - PubMed Abstract:
Plasminogen is the proenzyme precursor of the primary fibrinolytic protease plasmin. Circulating plasminogen, which comprises a Pan-apple (PAp) domain, five kringle domains (KR1-5), and a serine protease (SP) domain, adopts a closed, activation-resistant conformation. The kringle domains mediate interactions with fibrin clots and cell-surface receptors. These interactions trigger plasminogen to adopt an open form that can be cleaved and converted to plasmin by tissue-type and urokinase-type plasminogen activators. Here, the structure of closed plasminogen reveals that the PAp and SP domains, together with chloride ions, maintain the closed conformation through interactions with the kringle array. Differences in glycosylation alter the position of KR3, although in all structures the loop cleaved by plasminogen activators is inaccessible. The ligand-binding site of KR1 is exposed and likely governs proenzyme recruitment to targets. Furthermore, analysis of our structure suggests that KR5 peeling away from the PAp domain may initiate plasminogen conformational change.
- Department of Biochemistry and Molecular Biology, Monash University, Clayton, Melbourne, VIC 3800 Australia.
Organizational Affiliation: