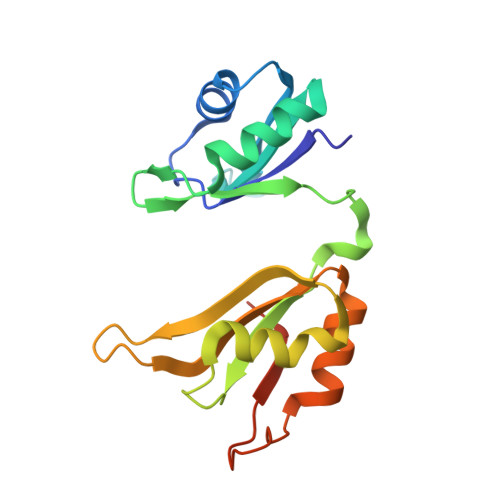
The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding.
Wang, H., Zeng, F., Liu, Q., Liu, H., Liu, Z., Niu, L., Teng, M., Li, X.(2013) Acta Crystallogr D Biol Crystallogr 69: 373-380
- PubMed: 23519412
- DOI: https://doi.org/10.1107/S0907444912047828
- Primary Citation of Related Structures:
4ED5 - PubMed Abstract:
Human RNA-binding protein (HuR), a ubiquitously expressed member of the Hu protein family, plays an important role in mRNA degradation and has been implicated as a key post-transcriptional regulator. HuR contains three RNA-recognition motif (RRM) domains. The two N-terminal tandem RRM domains can selectively bind AU-rich elements (AREs), while the third RRM domain (RRM3) contributes to interactions with the poly-A tail of target mRNA and other ligands. Here, the X-ray structure of two methylated tandem RRM domains (RRM1/2) of HuR in their RNA-free form was solved at 2.9 Å resolution. The crystal structure of RRM1/2 complexed with target mRNA was also solved at 2.0 Å resolution; comparisons of the two structures show that HuR RRM1/2 undergoes conformational changes upon RNA binding. Fluorescence polarization assays (FPA) were used to study the protein-RNA interactions. Both the structure and the FPA analysis indicated that RRM1 is the primary ARE-binding domain in HuR and that the conformational changes induce subsequent contacts of the RNA substrate with the inter-domain linker and RRM2 which greatly improve the RNA-binding affinity of HuR.
- School of Life Sciences, University of Science and Technology of China, 96 Jinzhai Road, Hefei, Anhui 230026, People's Republic of China.
Organizational Affiliation: