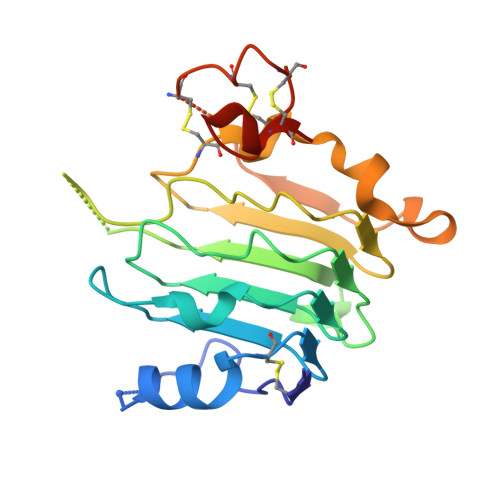
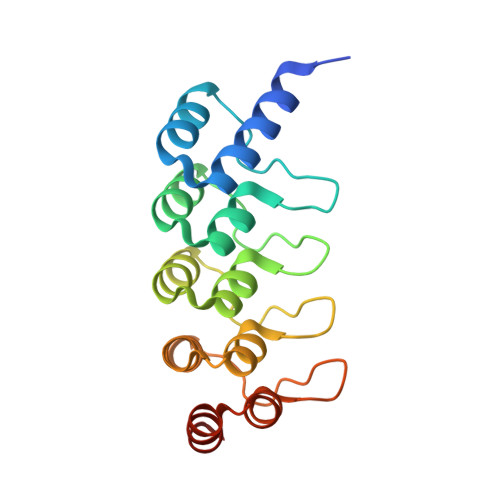
Structural Basis for Eliciting a Cytotoxic Effect in HER2-Overexpressing Cancer Cells via Binding to the Extracellular Domain of HER2.
Jost, C., Schilling, J., Tamaskovic, R., Schwill, M., Honegger, A., Plueckthun, A.(2013) Structure 21: 1-13
- PubMed: 24095059
- DOI: https://doi.org/10.1016/j.str.2013.08.020
- Primary Citation of Related Structures:
4HRL, 4HRM, 4HRN - PubMed Abstract:
Human epidermal growth factor receptor-2 (HER2) is a receptor tyrosine kinase directly linked to the growth of malignancies from various origins and a validated target for monoclonal antibodies and kinase inhibitors. Utilizing a new approach with designed ankyrin repeat proteins (DARPins) as alternative binders, we show that binding of two DARPins connected by a short linker, one targeting extracellular subdomain I and the other subdomain IV, causes much stronger cytotoxic effects on the HER2-addicted breast cancer cell line BT474, surpassing the therapeutic antibody trastuzumab. We determined crystal structures of these DARPins in complex with the respective subdomains. Detailed models of the full-length receptor, constrained by its rigid domain structures and its membrane anchoring, explain how the bispecific DARPins connect two membrane-bound HER2 molecules, distorting them such that they cannot form signaling-competent dimers with any EGFR family member, preventing any kinase dimerization, and thus leading to a complete loss of signaling.
- Department of Biochemistry, University of Zürich, 8057 Zürich, Switzerland.
Organizational Affiliation: