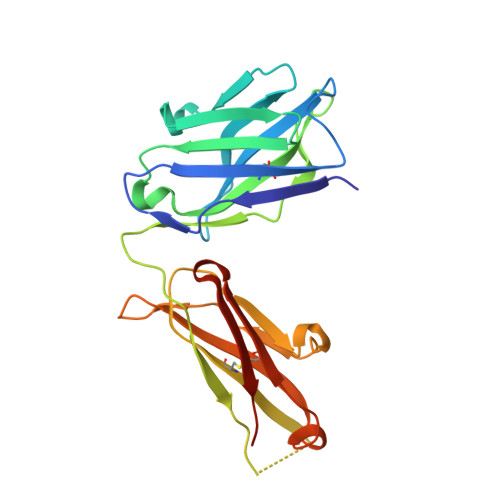
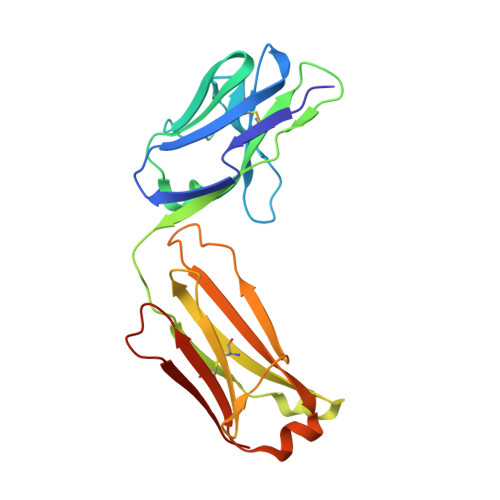
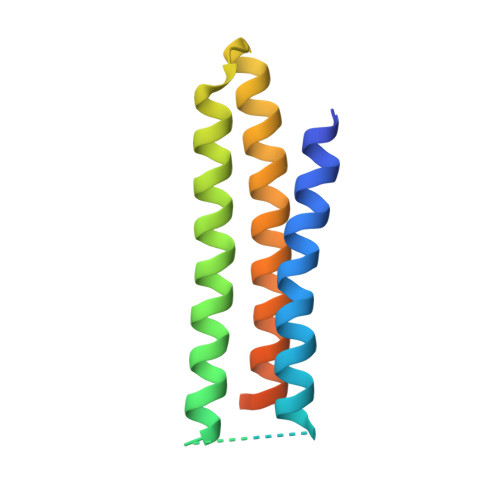
Proof of principle for epitope-focused vaccine design.
Correia, B.E., Bates, J.T., Loomis, R.J., Baneyx, G., Carrico, C., Jardine, J.G., Rupert, P., Correnti, C., Kalyuzhniy, O., Vittal, V., Connell, M.J., Stevens, E., Schroeter, A., Chen, M., Macpherson, S., Serra, A.M., Adachi, Y., Holmes, M.A., Li, Y., Klevit, R.E., Graham, B.S., Wyatt, R.T., Baker, D., Strong, R.K., Crowe, J.E., Johnson, P.R., Schief, W.R.(2014) Nature 507: 201-206
- PubMed: 24499818
- DOI: https://doi.org/10.1038/nature12966
- Primary Citation of Related Structures:
4JLR, 4L8I, 4N9G - PubMed Abstract:
Vaccines prevent infectious disease largely by inducing protective neutralizing antibodies against vulnerable epitopes. Several major pathogens have resisted traditional vaccine development, although vulnerable epitopes targeted by neutralizing antibodies have been identified for several such cases. Hence, new vaccine design methods to induce epitope-specific neutralizing antibodies are needed. Here we show, with a neutralization epitope from respiratory syncytial virus, that computational protein design can generate small, thermally and conformationally stable protein scaffolds that accurately mimic the viral epitope structure and induce potent neutralizing antibodies. These scaffolds represent promising leads for the research and development of a human respiratory syncytial virus vaccine needed to protect infants, young children and the elderly. More generally, the results provide proof of principle for epitope-focused and scaffold-based vaccine design, and encourage the evaluation and further development of these strategies for a variety of other vaccine targets, including antigenically highly variable pathogens such as human immunodeficiency virus and influenza.
- 1] Department of Biochemistry, University of Washington, Seattle, Washington 98195, USA [2] PhD Program in Computational Biology, Instituto Gulbenkian Ciência and Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Oeiras 2780-157, Portugal [3] Department of Chemical Physiology, The Scripps Research Institute, La Jolla, California 92037, USA.
Organizational Affiliation: