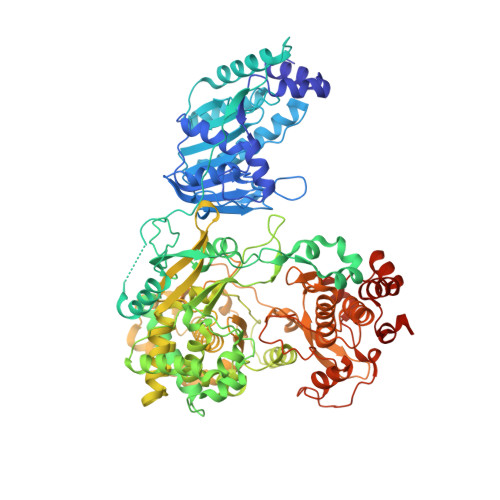
Crystal Structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface
Lu, G., Gong, P.(2013) PLoS Pathog 9: e1003549-e1003549
- PubMed: 23950717
- DOI: https://doi.org/10.1371/journal.ppat.1003549
- Primary Citation of Related Structures:
4K6M - PubMed Abstract:
The flavivirus NS5 harbors a methyltransferase (MTase) in its N-terminal ≈ 265 residues and an RNA-dependent RNA polymerase (RdRP) within the C-terminal part. One of the major interests and challenges in NS5 is to understand the interplay between RdRP and MTase as a unique natural fusion protein in viral genome replication and cap formation. Here, we report the first crystal structure of the full-length flavivirus NS5 from Japanese encephalitis virus. The structure completes the vision for polymerase motifs F and G, and depicts defined intra-molecular interactions between RdRP and MTase. Key hydrophobic residues in the RdRP-MTase interface are highly conserved in flaviviruses, indicating the biological relevance of the observed conformation. Our work paves the way for further dissection of the inter-regulations of the essential enzymatic activities of NS5 and exploration of possible other conformations of NS5 under different circumstances.
- State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuchang District, Wuhan, Hubei, China.
Organizational Affiliation: