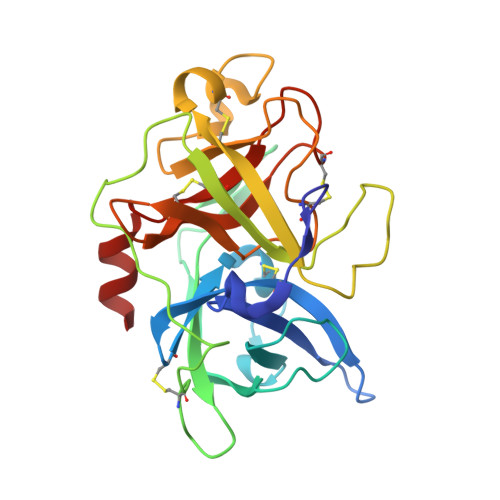
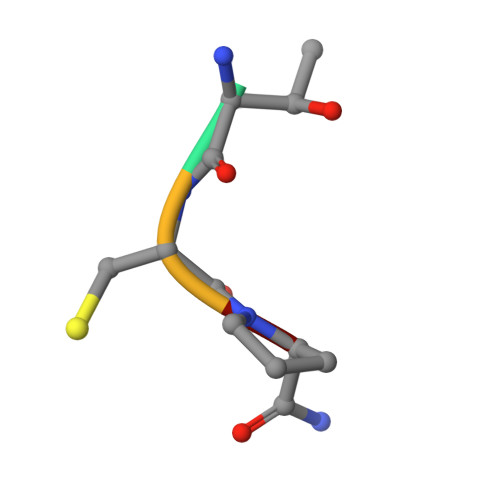
Peptide ligands stabilized by small molecules.
Chen, S., Bertoldo, D., Angelini, A., Pojer, F., Heinis, C.(2014) Angew Chem Int Ed Engl 53: 1602-1606
- PubMed: 24453110
- DOI: https://doi.org/10.1002/anie.201309459
- Primary Citation of Related Structures:
4MNV, 4MNW, 4MNX, 4MNY - PubMed Abstract:
Bicyclic peptides generated through directed evolution by using phage display offer an attractive ligand format for the development of therapeutics. Being nearly 100-fold smaller than antibodies, they promise advantages such as access to chemical synthesis, efficient diffusion into tissues, and needle-free application. However, unlike antibodies, they do not have a folded structure in solution and thus bind less well. We developed bicyclic peptides with hydrophilic chemical structures at their center to promote noncovalent intramolecular interactions, thereby stabilizing the peptide conformation. The sequences of the peptides isolated by phage display from large combinatorial libraries were strongly influenced by the type of small molecule used in the screen, thus suggesting that the peptides fold around the small molecules. X-ray structure analysis revealed that the small molecules indeed formed hydrogen bonds with the peptides. These noncovalent interactions stabilize the peptide-protein complexes and contribute to the high binding affinity.
- Institute of Chemical Sciences and Engineering, Ecole Polytechnique Fédérale de Lausanne, 1015 Lausanne (Switzerland).
Organizational Affiliation: