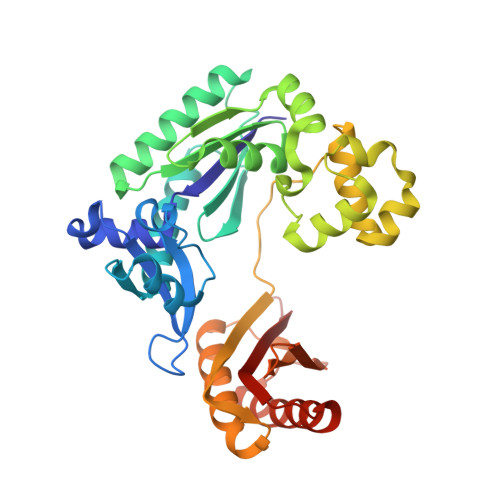
Structural and kinetic insights into binding and incorporation of L-nucleotide analogs by a Y-family DNA polymerase.
Gaur, V., Vyas, R., Fowler, J.D., Efthimiopoulos, G., Feng, J.Y., Suo, Z.(2014) Nucleic Acids Res 42: 9984-9995
- PubMed: 25104018
- DOI: https://doi.org/10.1093/nar/gku709
- Primary Citation of Related Structures:
4QW8, 4QW9, 4QWA, 4QWB, 4QWC, 4QWD, 4QWE - PubMed Abstract:
Considering that all natural nucleotides (D-dNTPs) and the building blocks (D-dNMPs) of DNA chains possess D-stereochemistry, DNA polymerases and reverse transcriptases (RTs) likely possess strongD-stereoselectivity by preferably binding and incorporating D-dNTPs over unnatural L-dNTPs during DNA synthesis. Surprisingly, a structural basis for the discrimination against L-dNTPs by DNA polymerases or RTs has not been established although L-deoxycytidine analogs (lamivudine and emtricitabine) and L-thymidine (telbivudine) have been widely used as antiviral drugs for years. Here we report seven high-resolution ternary crystal structures of a prototype Y-family DNA polymerase, DNA, and D-dCTP, D-dCDP, L-dCDP, or the diphosphates and triphosphates of lamivudine and emtricitabine. These structures reveal that relative to D-dCTP, each of these L-nucleotides has its sugar ring rotated by 180° with an unusual O4'-endo sugar puckering and exhibits multiple triphosphate-binding conformations within the active site of the polymerase. Such rare binding modes significantly decrease the incorporation rates and efficiencies of these L-nucleotides catalyzed by the polymerase.
- Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210, USA.
Organizational Affiliation: