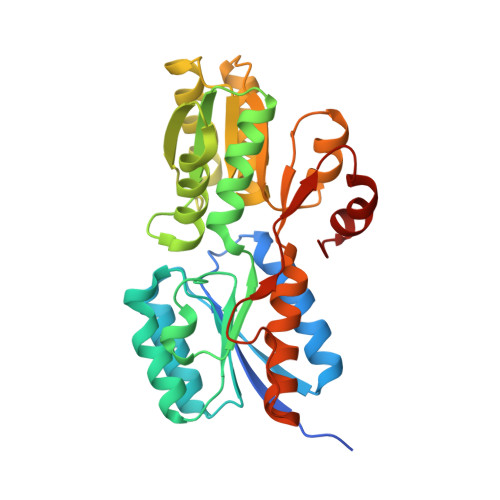
Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus, TARGET EFI-510965.
Patskovsky, Y., Toro, R., Bhosle, R., Al Obaidi, N., Chamala, S., Scott Glenn, A., Attonito, J.D., Chowdhury, S., Lafleur, J., Siedel, R.D., Morisco, L.L., Wasserman, S.R., Hillerich, B., Love, J., Whalen, K.L., Gerlt, J.A., Almo, S.C.To be published.