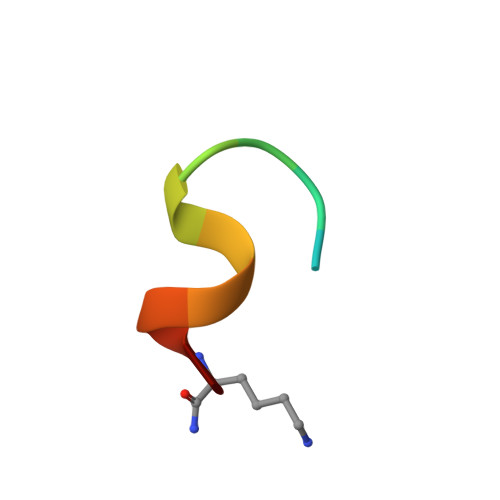Structural Determinants of Integrin Beta-Subunit Specificity for Latent Tgf-Beta
Dong, X., Hudson, N.E., Lu, C., Springer, T.A.(2014) Nat Struct Mol Biol 21: 1091
- PubMed: 25383667
- DOI: https://doi.org/10.1038/nsmb.2905
- Primary Citation of Related Structures:
4UM8, 4UM9 - PubMed Abstract:
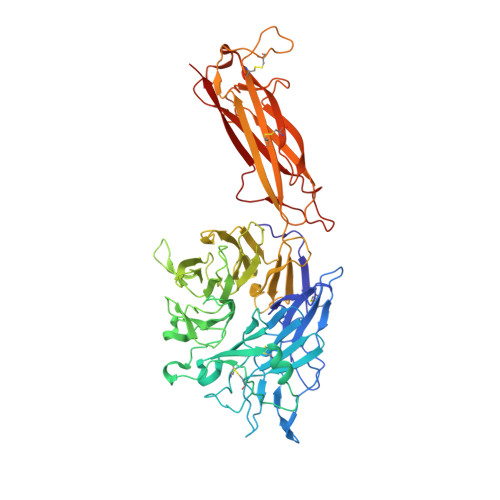
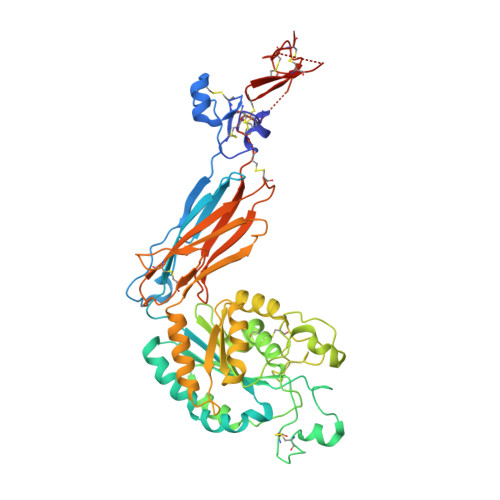
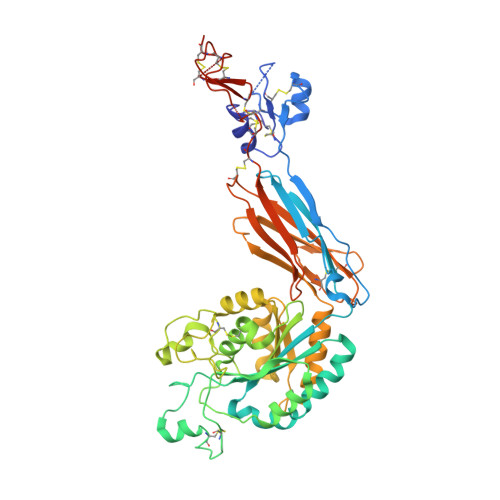
Eight integrin α-β heterodimers recognize ligands with an Arg-Gly-Asp (RGD) motif. However, the structural mechanism by which integrins differentiate among extracellular proteins with RGD motifs is not understood. Here, crystal structures, mutations and peptide-affinity measurements show that αVβ6 binds with high affinity to a RGDLXXL/I motif within the prodomains of TGF-β1 and TGF-β3. The LXXL/I motif forms an amphipathic α-helix that binds in a hydrophobic pocket in the β6 subunit. Elucidation of the basis for ligand binding specificity by the integrin β subunit reveals contributions by three different βI-domain loops, which we designate specificity-determining loops (SDLs) 1, 2 and 3. Variation in a pair of single key residues in SDL1 and SDL3 correlates with the variation of the entire β subunit in integrin evolution, thus suggesting a paradigmatic role in overall β-subunit function.
- 1] Boston Children's Hospital, Harvard Medical School, Boston, Massachusetts, USA. [2] Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts, USA.
Organizational Affiliation: