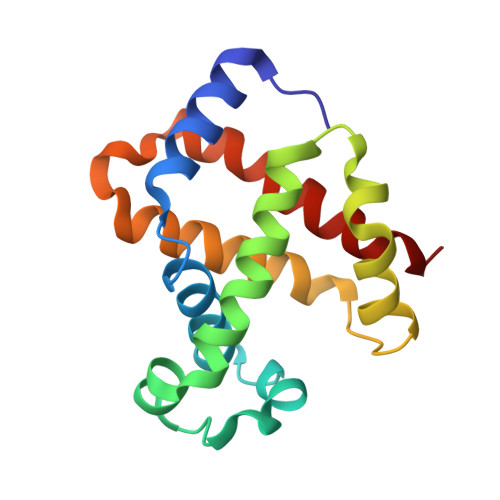
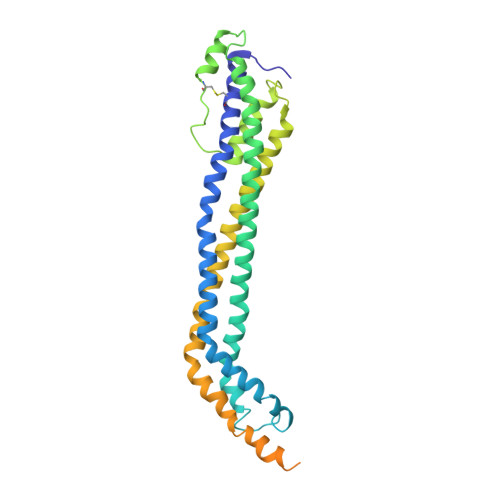
Structural basis for trypanosomal haem acquisition and susceptibility to the host innate immune system.
Stdkilde, K., Torvund-Jensen, M., Moestrup, S.K., Andersen, C.B.(2014) Nat Commun 5: 5487-5487
- PubMed: 25410714
- DOI: https://doi.org/10.1038/ncomms6487
- Primary Citation of Related Structures:
4WJG - PubMed Abstract:
Sleeping sickness is caused by trypanosome parasites, which infect humans and livestock in Sub-Saharan Africa. Haem is an important growth factor for the parasites and is acquired from the host by receptor-mediated uptake of haptoglobin (Hp)-haemoglobin (Hb) complexes. The parasite Hp-Hb receptor (HpHbR) is also a target for a specialized innate immune defence executed by trypanosome-killing lipoprotein particles containing an Hp-related protein in complex with Hb. Here we report the structure of the multimeric complex between human Hp-Hb and Trypanosoma brucei brucei HpHbR. Two receptors forming kinked three-helical rods with small head regions bind to Hp and the β-subunits of Hb (βHb), with one receptor at each end of the dimeric Hp-Hb complex. The Hb β-subunit haem group directly associates with the receptors, which allows for sensing of haem-containing Hp-Hb. The HpHbR-binding region of Hp is conserved in Hp-related protein, indicating an identical recognition of Hp-Hb and trypanolytic particles by HpHbR in human plasma.
- Department of Biomedicine, Aarhus University, Ole Worms Allé 3, DK-8000 Aarhus C, Denmark.
Organizational Affiliation: