Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4
Sugahara, M., Ago, H., Saino, H., Miyano, M., Yamamoto, S., Takahashi, Y.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| anti-leukotriene C4 monoclonal antibody immunoglobulin kappa light chain | 238 | Mus musculus | Mutation(s): 0 | 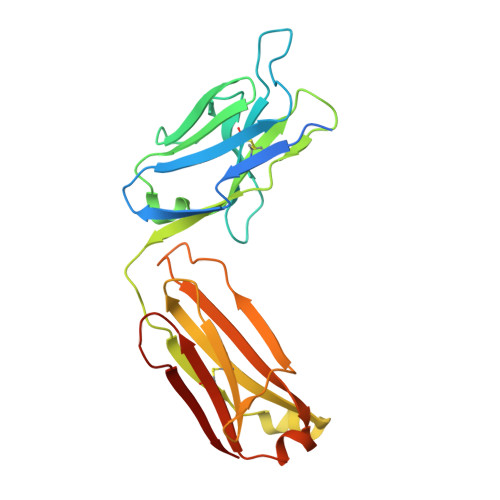 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| anti-leukotriene C4 monoclonal antibody immunoglobulin gamma1 heavy chain | 237 | Mus musculus | Mutation(s): 0 | 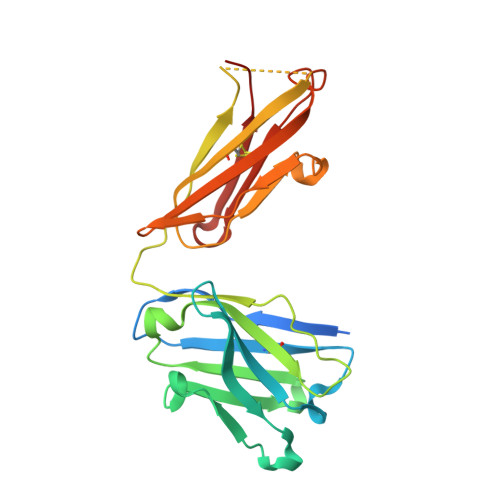 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LTX Query on LTX | I [auth A], L [auth C], P [auth E], T [auth G] | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-(
2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid C30 H47 N3 O9 S GWNVDXQDILPJIG-NXOLIXFESA-N |  | ||
| MES Query on MES | K [auth B], O [auth D], R [auth F], V [auth H] | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID C6 H13 N O4 S SXGZJKUKBWWHRA-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | J [auth B] M [auth C] N [auth D] Q [auth E] S [auth G] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 74.6 | α = 90 |
| b = 173.98 | β = 90.63 |
| c = 75.46 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| CNS | refinement |
| REFMAC | refinement |
| iMOSFLM | data processing |
| SCALA | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Education, Culture, Sports, Science and Technology | Japan | S1311005 |
| RIKEN | Japan | -- |