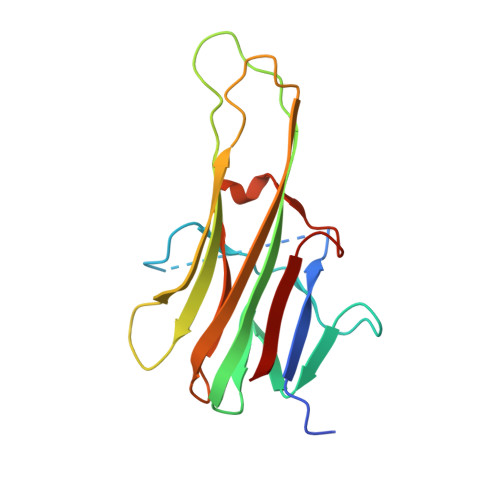
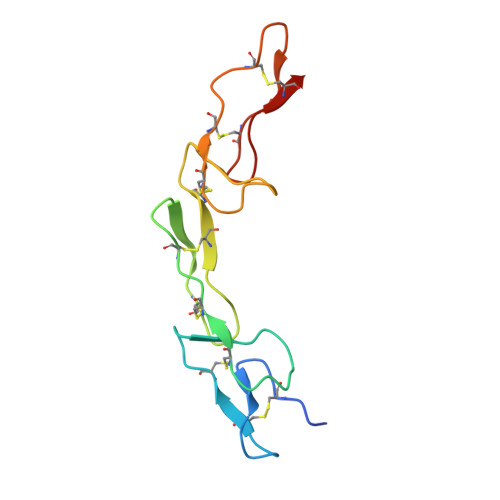
The structure of the death receptor 4-TNF-related apoptosis-inducing ligand (DR4-TRAIL) complex.
Ramamurthy, V., Yamniuk, A.P., Lawrence, E.J., Yong, W., Schneeweis, L.A., Cheng, L., Murdock, M., Corbett, M.J., Doyle, M.L., Sheriff, S.(2015) Acta Crystallogr F Struct Biol Commun 71: 1273-1281
- PubMed: 26457518
- DOI: https://doi.org/10.1107/S2053230X15016416
- Primary Citation of Related Structures:
5CIR - PubMed Abstract:
The structure of death receptor 4 (DR4) in complex with TNF-related apoptosis-inducing ligand (TRAIL) has been determined at 3 Å resolution and compared with those of previously determined DR5-TRAIL complexes. Consistent with the high sequence similarity between DR4 and DR5, the overall arrangement of the DR4-TRAIL complex does not differ substantially from that of the DR5-TRAIL complex. However, subtle differences are apparent. In addition, solution interaction studies were carried out that show differences in the thermodynamics of binding DR4 or DR5 with TRAIL.
- Molecular Structure and Design, Bristol-Myers Squibb R&D, PO Box 4000, Princeton, NJ 08543-4000, USA.
Organizational Affiliation: