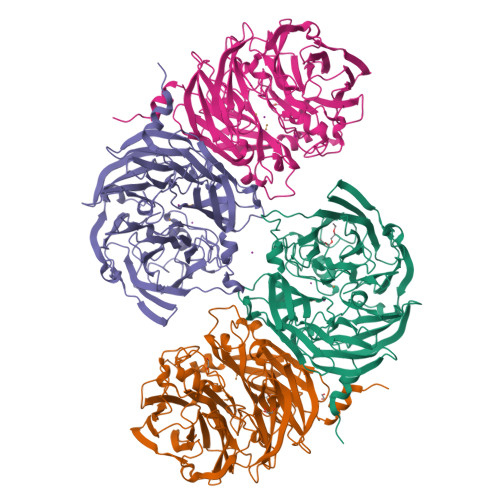
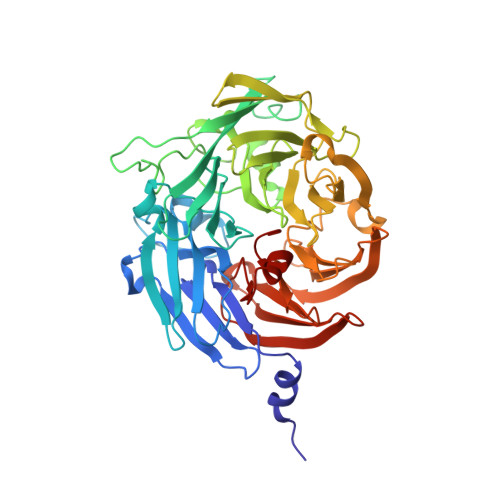
Discovery and characterization of a novel copper containing enzyme - THIOCYANATE DEHYDROGENASE.
Tikhonova, T.V., Sorokin, D.I., Tsallagov, S.I., Polyakov, K.M., Trofimov, A.A., Shabalin, I.G., Khrenova, M.G., Muyser, G., Rakitina, T.V., Popov, V.O.To be published.