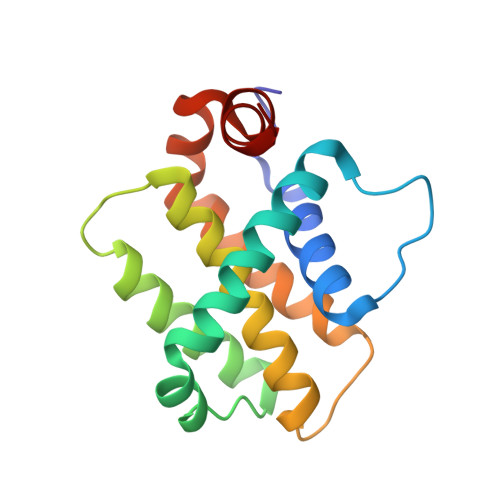
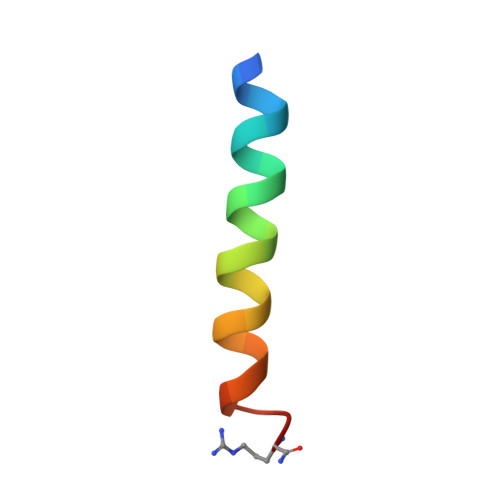
Epistatic mutations in PUMA BH3 drive an alternate binding mode to potently and selectively inhibit anti-apoptotic Bfl-1.
Jenson, J.M., Ryan, J.A., Grant, R.A., Letai, A., Keating, A.E.(2017) Elife 6
- PubMed: 28594323
- DOI: https://doi.org/10.7554/eLife.25541
- Primary Citation of Related Structures:
5UUK, 5UUL, 5UUM, 5UUP - PubMed Abstract:
Overexpression of anti-apoptotic Bcl-2 family proteins contributes to cancer progression and confers resistance to chemotherapy. Small molecules that target Bcl-2 are used in the clinic to treat leukemia, but tight and selective inhibitors are not available for Bcl-2 paralog Bfl-1. Guided by computational analysis, we designed variants of the native BH3 motif PUMA that are > 150-fold selective for Bfl-1 binding. The designed peptides potently trigger disruption of the mitochondrial outer membrane in cells dependent on Bfl-1, but not in cells dependent on other anti-apoptotic homologs. High-resolution crystal structures show that designed peptide FS2 binds Bfl-1 in a shifted geometry, relative to PUMA and other binding partners, due to a set of epistatic mutations. FS2 modified with an electrophile reacts with a cysteine near the peptide-binding groove to augment specificity. Designed Bfl-1 binders provide reagents for cellular profiling and leads for developing enhanced and cell-permeable peptide or small-molecule inhibitors.
- Department of Biology, Massachusetts Institute of Technology, Cambridge, United States.
Organizational Affiliation: