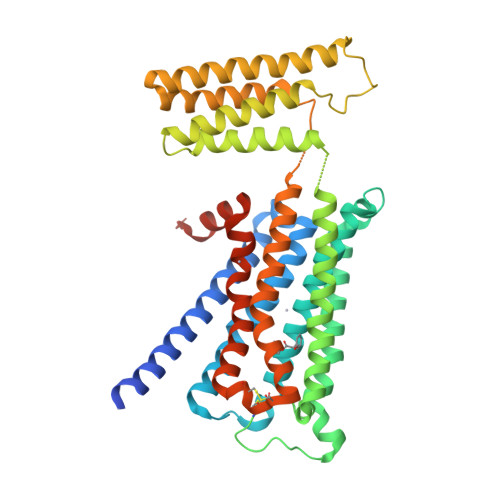
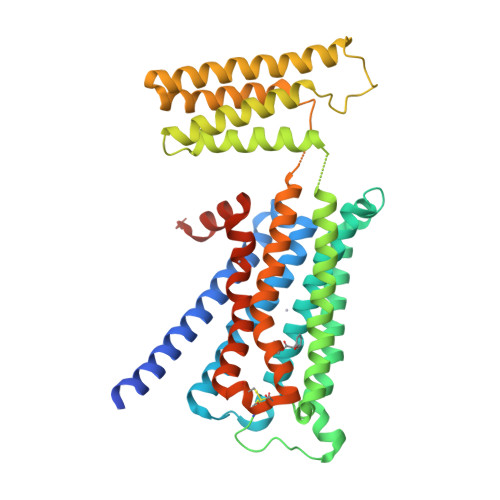
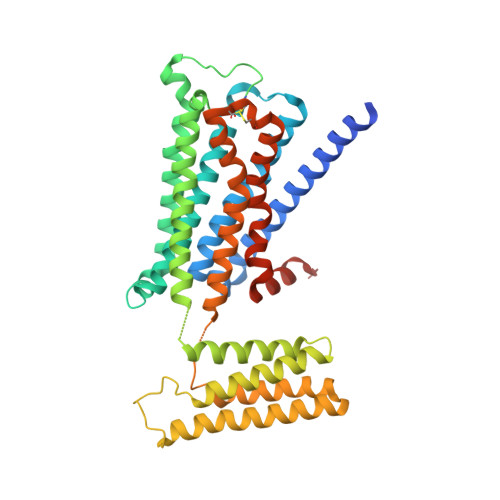
Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Suno, R., Lee, S., Maeda, S., Yasuda, S., Yamashita, K., Hirata, K., Horita, S., Tawaramoto, M.S., Tsujimoto, H., Murata, T., Kinoshita, M., Yamamoto, M., Kobilka, B.K., Vaidehi, N., Iwata, S., Kobayashi, T.(2018) Nat Chem Biol 14: 1150-1158
- PubMed: 30420692
- DOI: https://doi.org/10.1038/s41589-018-0152-y
- Primary Citation of Related Structures:
5YC8, 5ZK3, 5ZK8, 5ZKB, 5ZKC - PubMed Abstract:
Human muscarinic receptor M 2 is one of the five subtypes of muscarinic receptors belonging to the family of G-protein-coupled receptors. Muscarinic receptors are targets for multiple neurodegenerative diseases. The challenge has been designing subtype-selective ligands against one of the five muscarinic receptors. We report high-resolution structures of a thermostabilized mutant M 2 receptor bound to a subtype-selective antagonist AF-DX 384 and a nonselective antagonist NMS. The thermostabilizing mutation S110R in M 2 was predicted using a theoretical strategy previously developed in our group. Comparison of the crystal structures and pharmacological properties of the M 2 receptor shows that the Arg in the S110R mutant mimics the stabilizing role of the sodium cation, which is known to allosterically stabilize inactive state(s) of class A GPCRs. Molecular dynamics simulations reveal that tightening of the ligand-residue contacts in M 2 receptors compared to M 3 receptors leads to subtype selectivity of AF-DX 384.
Organizational Affiliation:
Department of Cell Biology and Medical Chemistry, Graduate School of Medicine, Kyoto University, Konoe-cho, Yoshida, Sakyo-ku, Kyoto, Japan. r.suno@mfour.med.kyoto-u.ac.jp.