An Effective Neutralizing Antibody Against Influenza Virus H1N1 from Human B Cells.
Lee, C.C., Yang, C.Y., Lin, L.L., Ko, T.P., Chang, A.H., Chang, S.S., Wang, A.H.(2019) Sci Rep 9: 4546-4546
- PubMed: 30872685
- DOI: https://doi.org/10.1038/s41598-019-40937-4
- Primary Citation of Related Structures:
6A4K - PubMed Abstract:
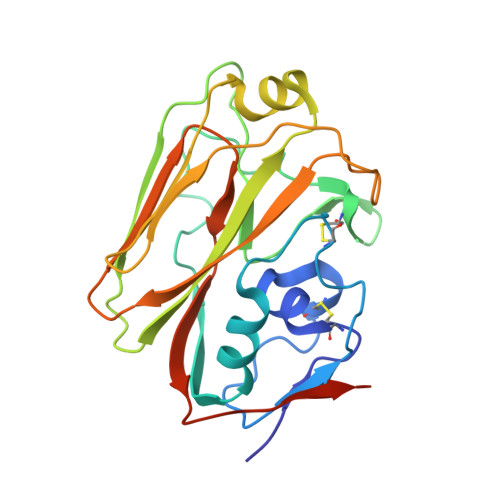
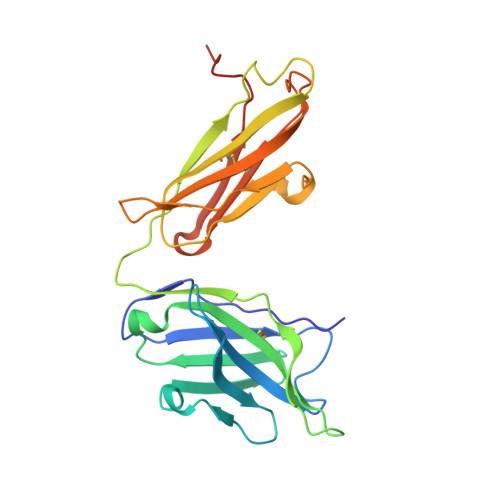
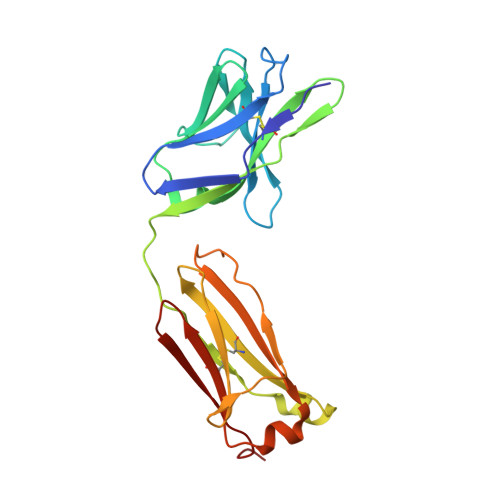
Influenza is a contagious acute respiratory disease caused by the influenza virus infection. Hemagglutinin (HA) is an important target in the therapeutic treatment and diagnostic detection of the influenza virus. Influenza A virus encompasses several different HA subtypes with different strains, which are constantly changing. In this study, we identified a fully human H1N1 neutralizing antibody (32D6) via an Epstein-Barr virus-immortalized B cell-based technology. 32D6 specifically neutralizes the clinically isolated H1N1 strains after the 2009 pandemic but not the earlier strains. The epitope was identified through X-ray crystallographic analysis of the 32D6-Fab/HA1 complex structure, which revealed a unique loop conformation located on the top surface of HA. The major region is composed of two peptide segments (residues 172-177 and 206-213), which form an abreast loop conformation. The residue T262 between the two loops forms a conformational epitope for recognition by 32D6. Three water molecules were observed at the interface of HA and the heavy chain, and they may constitute a stabilizing element for the 32D6-HA association. In addition, each 32D6-Fab is likely capable of blocking one HA trimer. This study provides important information on the strain specificity of 32D6 for the therapeutic treatment and detection of viral infection.
- Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan.
Organizational Affiliation: