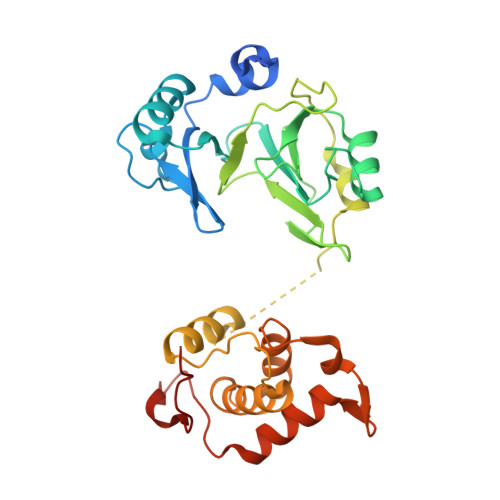
Crystal structure and biochemical studies of the bifunctional DNA primase-polymerase from phage NrS-1.
Guo, H.J., Li, M.J., Wang, T.L., Wu, H., Zhou, H., Xu, C.Y., Liu, X.P., Yu, F., He, J.H.(2019) Biochem Biophys Res Commun 510: 573-579
- PubMed: 30739783
- DOI: https://doi.org/10.1016/j.bbrc.2019.01.144
- Primary Citation of Related Structures:
6A9W - PubMed Abstract:
A novel DNA polymerase found in the deep-sea vent phage NrS-1, was confirmed to have both DNA polymerase and primase activities. In this polymerase, the N-terminal residues 1-300 (referred to as N300) are the core region required for polymerizing DNA and catalyzing de novo DNA synthesis. Here, the crystal structure of N300 was solved at a resolution of 1.80 Å. The overall structure consists of a prim/pol domain and a helix bundle domain, which are separated by a 14-residue-long flexible tether (residues 177-190). Both the prim/pol domain of N300 and other primase-polymerases (prim-pol) encompass an analogous fold with conserved catalytic residues. Mutagenesis and enzymatic activity assays show that the acidic active-site residue E139 is required for both polymerase and primase activities. Functional assays confirm the essentiality of the helix bundle domain for primase activity. Furthermore, we identified a mutant (N300-Y261A) of the helix bundle domain, which probably plays an indispensable role in the primer initiation and recognition of template DNA.
Organizational Affiliation:
Shanghai Institute of Applied Physics, Chinese Academy of Sciences, Shanghai, 201800, China; University of Chinese Academy of Sciences, Beijing, 100049, China.