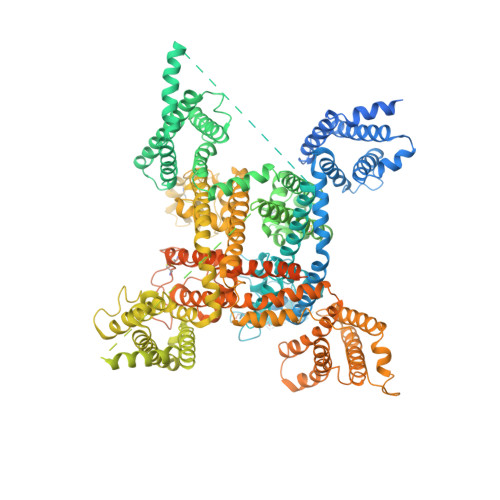
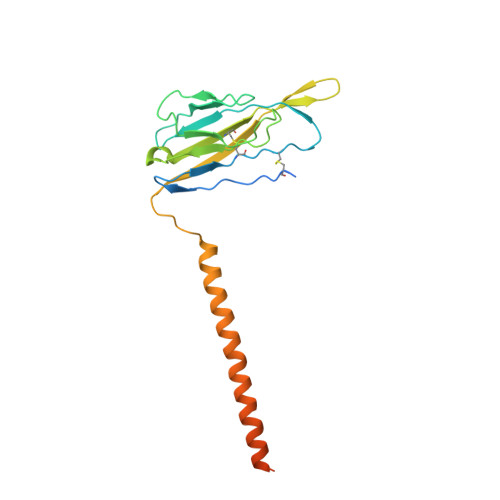
Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta 1.
Pan, X., Li, Z., Zhou, Q., Shen, H., Wu, K., Huang, X., Chen, J., Zhang, J., Zhu, X., Lei, J., Xiong, W., Gong, H., Xiao, B., Yan, N.(2018) Science 362
- PubMed: 30190309
- DOI: https://doi.org/10.1126/science.aau2486
- Primary Citation of Related Structures:
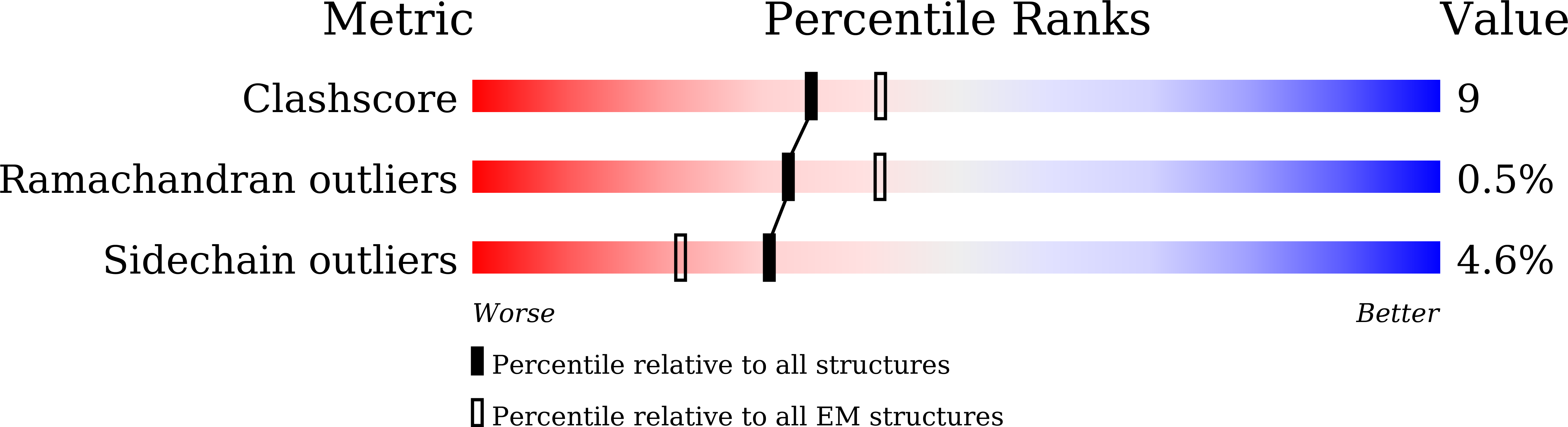
6AGF - PubMed Abstract:
Voltage-gated sodium (Na v ) channels, which are responsible for action potential generation, are implicated in many human diseases. Despite decades of rigorous characterization, the lack of a structure of any human Na v channel has hampered mechanistic understanding. Here, we report the cryo-electron microscopy structure of the human Na v 1.4-β1 complex at 3.2-Å resolution. Accurate model building was made for the pore domain, the voltage-sensing domains, and the β1 subunit, providing insight into the molecular basis for Na + permeation and kinetic asymmetry of the four repeats. Structural analysis of reported functional residues and disease mutations corroborates an allosteric blocking mechanism for fast inactivation of Na v channels. The structure provides a path toward mechanistic investigation of Na v channels and drug discovery for Na v channelopathies.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, Tsinghua University, Beijing 100084, China.