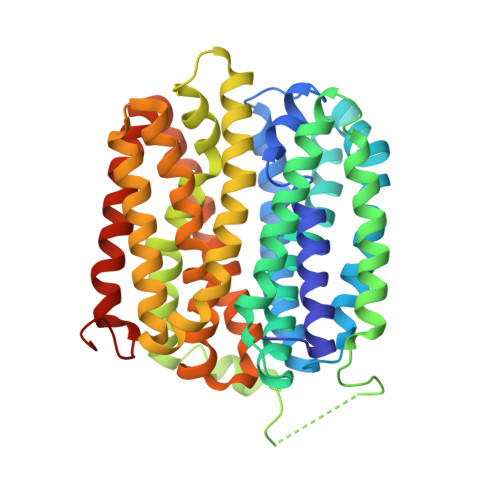
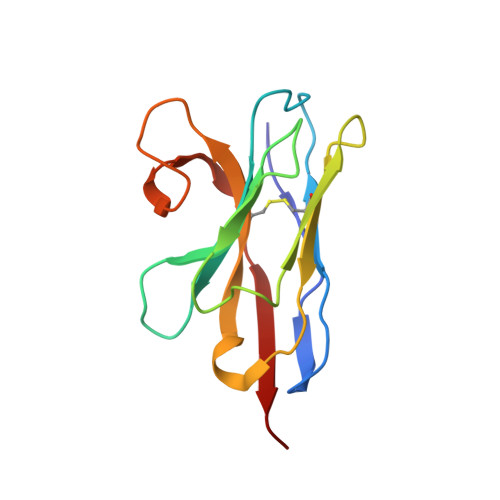
Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Kumar, H., Finer-Moore, J.S., Jiang, X., Smirnova, I., Kasho, V., Pardon, E., Steyaert, J., Kaback, H.R., Stroud, R.M.(2018) Proc Natl Acad Sci U S A 115: 8769-8774
- PubMed: 30108145
- DOI: https://doi.org/10.1073/pnas.1801774115
- Primary Citation of Related Structures:
6C9W - PubMed Abstract:
The lactose permease of Escherichia coli (LacY), a dynamic polytopic membrane transport protein, catalyzes galactoside/H + symport and operates by an alternating access mechanism that exhibits multiple conformations, the distribution of which is altered by sugar-binding. Camelid nanobodies were made against a double-mutant Gly46 → Trp/Gly262 → Trp (LacY WW ) that produces an outward-open conformation, as opposed to the cytoplasmic open-state crystal structure of WT LacY. Nanobody 9047 (Nb9047) stabilizes WT LacY in a periplasmic-open conformation. Here, we describe the X-ray crystal structure of a complex between LacY WW , the high-affinity substrate analog 4-nitrophenyl-α-d-galactoside (NPG), and Nb9047 at 3-Å resolution. The present crystal structure demonstrates that Nb9047 binds to the periplasmic face of LacY, primarily to the C-terminal six-helical bundle, while a flexible loop of the Nb forms a bridge between the N- and C-terminal halves of LacY across the periplasmic vestibule. The bound Nb partially covers the vestibule, yet does not affect the on-rates or off-rates for the substrate binding to LacY WW , which implicates dynamic flexibility of the Nb-LacY WW complex. Nb9047-binding neither changes the overall structure of LacY WW with bound NPG, nor the positions of side chains comprising the galactoside-binding site. The current NPG-bound structure exhibits a more occluded periplasmic vestibule than seen in a previous structure of a (different Nb) apo-LacY WW /Nb9039 complex that we argue is caused by sugar-binding, with major differences located at the periplasmic ends of transmembrane helices in the N-terminal half of LacY.
- Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94158.
Organizational Affiliation: