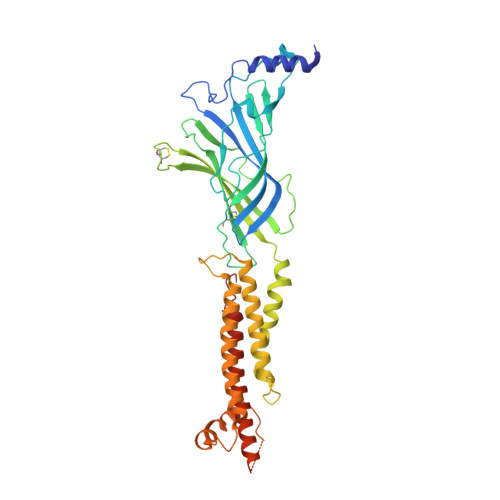
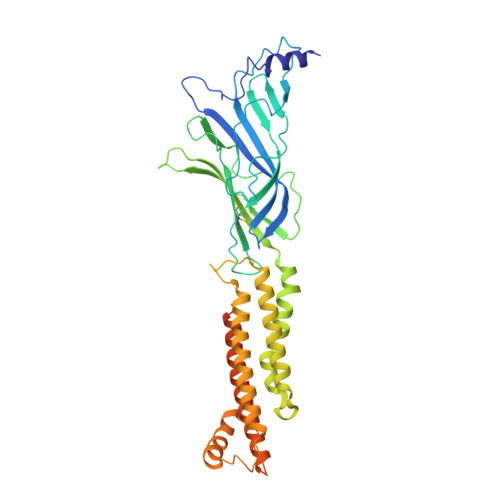
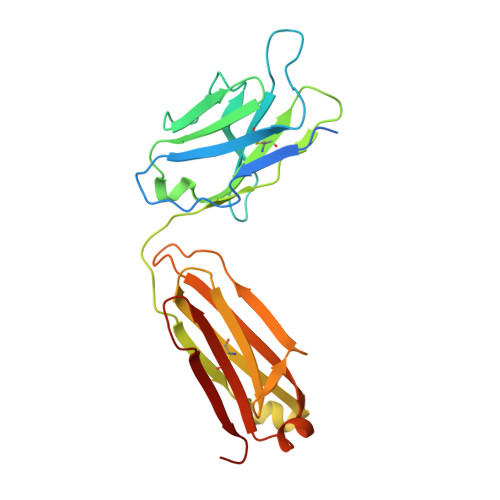
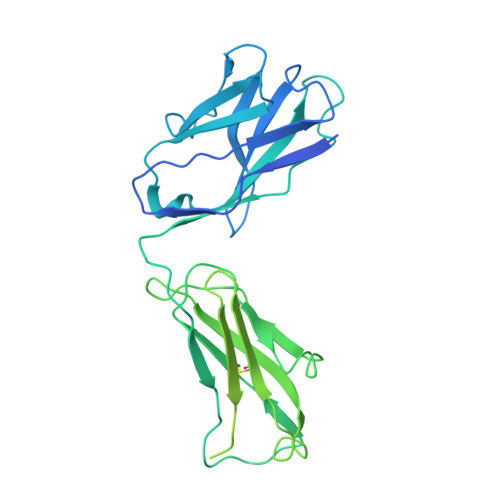
Structural principles of distinct assemblies of the human alpha 4 beta 2 nicotinic receptor.
Walsh, R.M., Roh, S.H., Gharpure, A., Morales-Perez, C.L., Teng, J., Hibbs, R.E.(2018) Nature 557: 261-265
- PubMed: 29720657
- DOI: https://doi.org/10.1038/s41586-018-0081-7
- Primary Citation of Related Structures:
6CNJ, 6CNK - PubMed Abstract:
Fast chemical communication in the nervous system is mediated by neurotransmitter-gated ion channels. The prototypical member of this class of cell surface receptors is the cation-selective nicotinic acetylcholine receptor. As with most ligand-gated ion channels, nicotinic receptors assemble as oligomers of subunits, usually as hetero-oligomers and often with variable stoichiometries 1 . This intrinsic heterogeneity in protein composition provides fine tunability in channel properties, which is essential to brain function, but frustrates structural and biophysical characterization. The α4β2 subtype of the nicotinic acetylcholine receptor is the most abundant isoform in the human brain and is the principal target in nicotine addiction. This pentameric ligand-gated ion channel assembles in two stoichiometries of α- and β-subunits (2α:3β and 3α:2β). Both assemblies are functional and have distinct biophysical properties, and an imbalance in the ratio of assemblies is linked to both nicotine addiction 2,3 and congenital epilepsy 4,5 . Here we leverage cryo-electron microscopy to obtain structures of both receptor assemblies from a single sample. Antibody fragments specific to β2 were used to 'break' symmetry during particle alignment and to obtain high-resolution reconstructions of receptors of both stoichiometries in complex with nicotine. The results reveal principles of subunit assembly and the structural basis of the distinctive biophysical and pharmacological properties of the two different stoichiometries of this receptor.
- Department of Neuroscience, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: