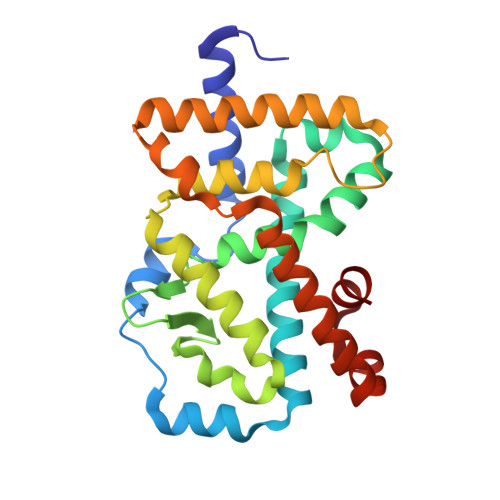
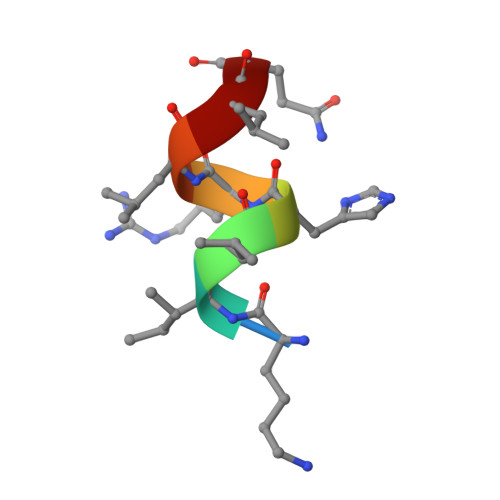
Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
Yukawa, T., Nara, Y., Kono, M., Sato, A., Oda, T., Takagi, T., Sato, T., Banno, Y., Taya, N., Imada, T., Shiokawa, Z., Negoro, N., Kawamoto, T., Koyama, R., Uchiyama, N., Skene, R., Hoffman, I., Chen, C.H., Sang, B., Snell, G., Katsuyama, R., Yamamoto, S., Shirai, J.(2019) J Med Chem 62: 1167-1179
- PubMed: 30652849
- DOI: https://doi.org/10.1021/acs.jmedchem.8b01181
- Primary Citation of Related Structures:
6E3E, 6E3G - PubMed Abstract:
Retinoic acid receptor-related orphan receptor γt (RORγt) agonists are expected to provide a novel class of immune-activating anticancer drugs via activation of Th17 cells and Tc17 cells. Herein, we describe a novel structure-based functionality switching approach from in house well-optimized RORγt inverse agonists to potent RORγt agonists. We succeeded in the identification of potent RORγt agonist 5 without major chemical structure change. The biochemical response was validated by molecular dynamics simulation studies that showed a helix 12 stabilization effect of RORγt agonists. These results indicate that targeting helix 12 is an attractive and novel medicinal chemistry strategy for switching existing RORγt inverse agonists to agonists.
- Pharmaceutical Research Division , Takeda Pharmaceutical Company Limited , 26-1 Muraoka-Higashi 2-chome , Fujisawa , Kanagawa 251-8555 , Japan.
Organizational Affiliation: