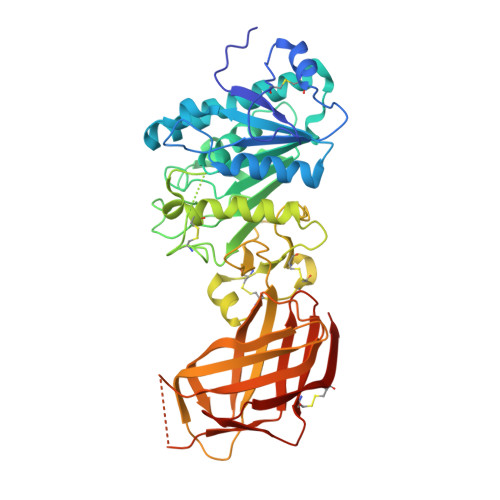
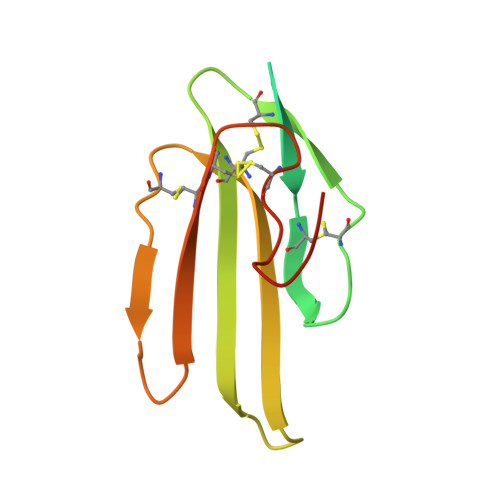
Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis.
Birrane, G., Beigneux, A.P., Dwyer, B., Strack-Logue, B., Kristensen, K.K., Francone, O.L., Fong, L.G., Mertens, H.D.T., Pan, C.Q., Ploug, M., Young, S.G., Meiyappan, M.(2019) Proc Natl Acad Sci U S A 116: 1723-1732
- PubMed: 30559189
- DOI: https://doi.org/10.1073/pnas.1817984116
- Primary Citation of Related Structures:
6E7K - PubMed Abstract:
Lipoprotein lipase (LPL) is responsible for the intravascular processing of triglyceride-rich lipoproteins. The LPL within capillaries is bound to GPIHBP1, an endothelial cell protein with a three-fingered LU domain and an N-terminal intrinsically disordered acidic domain. Loss-of-function mutations in LPL or GPIHBP1 cause severe hypertriglyceridemia (chylomicronemia), but structures for LPL and GPIHBP1 have remained elusive. Inspired by our recent discovery that GPIHBP1's acidic domain preserves LPL structure and activity, we crystallized an LPL-GPIHBP1 complex and solved its structure. GPIHBP1's LU domain binds to LPL's C-terminal domain, largely by hydrophobic interactions. Analysis of electrostatic surfaces revealed that LPL contains a large basic patch spanning its N- and C-terminal domains. GPIHBP1's acidic domain was not defined in the electron density map but was positioned to interact with LPL's large basic patch, providing a likely explanation for how GPIHBP1 stabilizes LPL. The LPL-GPIHBP1 structure provides insights into mutations causing chylomicronemia.
- Division of Experimental Medicine, Beth Israel Deaconess Medical Center, Boston, MA 02215.
Organizational Affiliation: