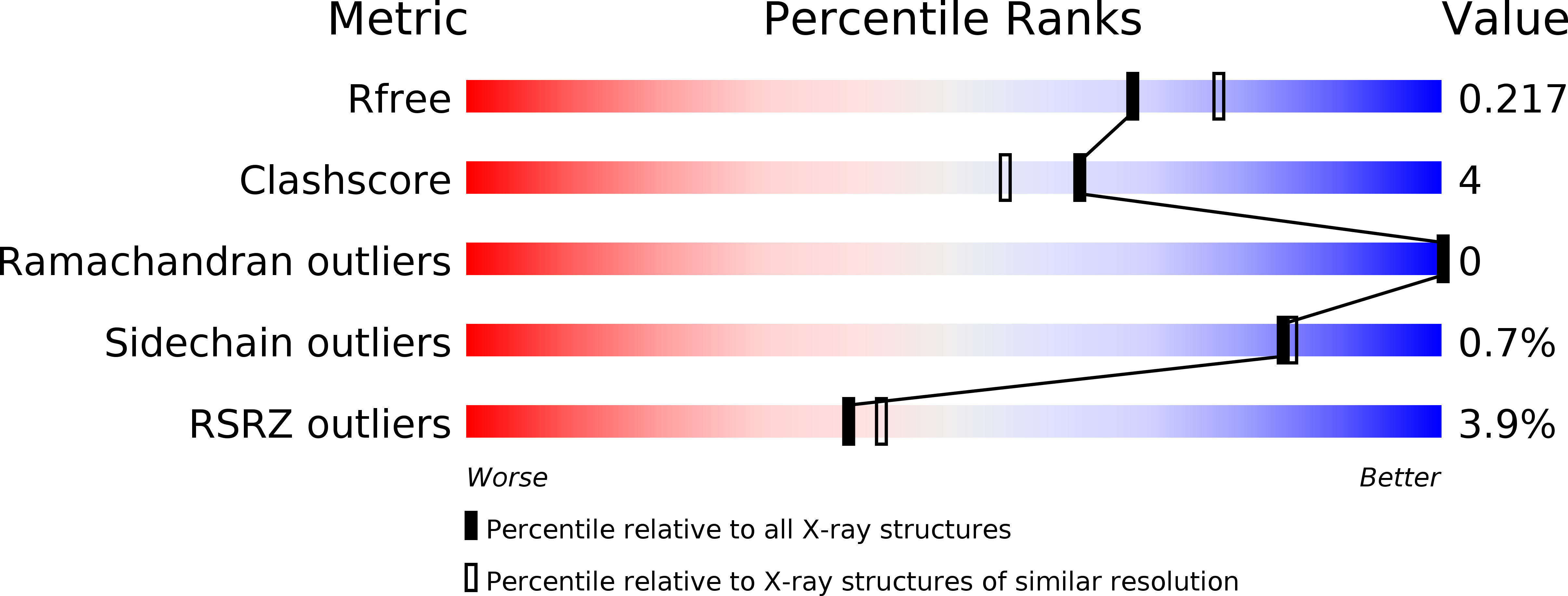
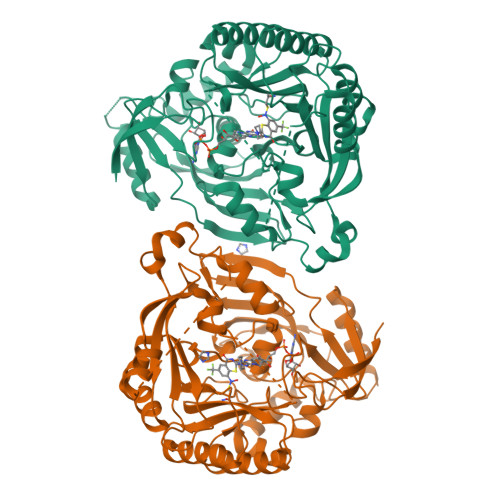
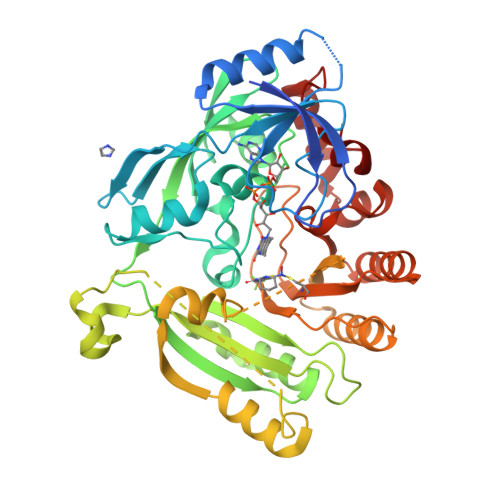
Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Richter, A., Rudolph, I., Mollmann, U., Voigt, K., Chung, C.W., Singh, O.M.P., Rees, M., Mendoza-Losana, A., Bates, R., Ballell, L., Batt, S., Veerapen, N., Futterer, K., Besra, G., Imming, P., Argyrou, A.(2018) Sci Rep 8: 13473-13473
- PubMed: 30194385
- DOI: https://doi.org/10.1038/s41598-018-31316-6
- Primary Citation of Related Structures:
6HEZ, 6HF0, 6HF3, 6HFV, 6HFW - PubMed Abstract:
Nitro-substituted 1,3-benzothiazinones (nitro-BTZs) are mechanism-based covalent inhibitors of Mycobacterium tuberculosis decaprenylphosphoryl-β-D-ribose-2'-oxidase (DprE1) with strong antimycobacterial properties. We prepared a number of oxidized and reduced forms of nitro-BTZs to probe the mechanism of inactivation of the enzyme and to identify opportunities for further chemistry. The kinetics of inactivation of DprE1 was examined using an enzymatic assay that monitored reaction progress up to 100 min, permitting compound ranking according to k inact /K i values. The side-chain at the 2-position and heteroatom identity at the 1-position of the BTZs were found to be important for inhibitory activity. We obtained crystal structures with several compounds covalently bound. The data suggest that steps upstream from the covalent end-points are likely the key determinants of potency and reactivity. The results of protein mass spectrometry using a 7-chloro-nitro-BTZ suggest that nucleophilic reactions at the 7-position do not operate and support a previously proposed mechanism in which BTZ activation by a reduced flavin intermediate is required. Unexpectedly, a hydroxylamino-BTZ showed time-dependent inhibition and mass spectrometry corroborated that this hydroxylamino-BTZ is a mechanism-based suicide inhibitor of DprE1. With this BTZ derivative, we propose a new covalent mechanism of inhibition of DprE1 that takes advantage of the oxidation cycle of the enzyme.
Organizational Affiliation:
Institut für Pharmazie, Martin-Luther-Universität Halle-Wittenberg, Wolfgang-Langenbeck-Str. 4, 06120, Halle (Saale), Germany.