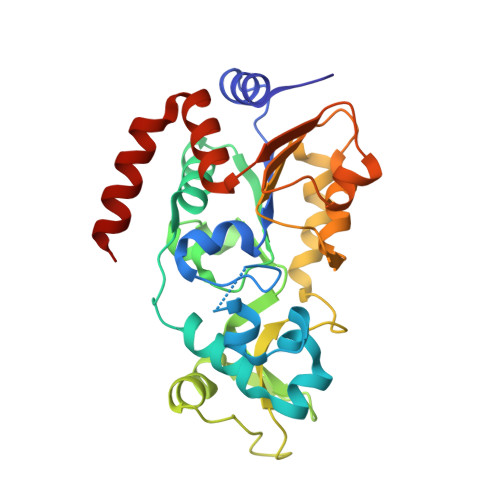
Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach.
Bao, X., Wang, Y., Li, X., Li, X.M., Liu, Z., Yang, T., Wong, C.F., Zhang, J., Hao, Q., Li, X.D.(2014) Elife 3
- PubMed: 25369635
- DOI: https://doi.org/10.7554/eLife.02999
- Primary Citation of Related Structures:
6ISO - PubMed Abstract:
Posttranslational modifications (PTMs) play a crucial role in a wide range of biological processes. Lysine crotonylation (Kcr) is a newly discovered histone PTM that is enriched at active gene promoters and potential enhancers in mammalian cell genomes. However, the cellular enzymes that regulate the addition and removal of Kcr are unknown, which has hindered further investigation of its cellular functions. Here we used a chemical proteomics approach to comprehensively profile 'eraser' enzymes that recognize a lysine-4 crotonylated histone H3 (H3K4Cr) mark. We found that Sirt1, Sirt2, and Sirt3 can catalyze the hydrolysis of lysine crotonylated histone peptides and proteins. More importantly, Sirt3 functions as a decrotonylase to regulate histone Kcr dynamics and gene transcription in living cells. This discovery not only opens opportunities for examining the physiological significance of histone Kcr, but also helps to unravel the unknown cellular mechanisms controlled by Sirt3, that have previously been considered solely as a deacetylase.
- Department of Chemistry, University of Hong Kong, Hong Kong, Hong Kong.
Organizational Affiliation: