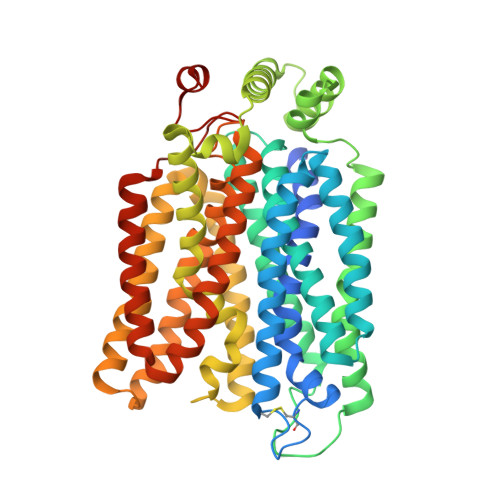
Structural Basis for Blocking Sugar Uptake into the Malaria Parasite Plasmodium falciparum.
Jiang, X., Yuan, Y., Huang, J., Zhang, S., Luo, S., Wang, N., Pu, D., Zhao, N., Tang, Q., Hirata, K., Yang, X., Jiao, Y., Sakata-Kato, T., Wu, J.W., Yan, C., Kato, N., Yin, H., Yan, N.(2020) Cell 183: 258-268.e12
- PubMed: 32860739
- DOI: https://doi.org/10.1016/j.cell.2020.08.015
- Primary Citation of Related Structures:
6M20, 6M2L - PubMed Abstract:
Plasmodium species, the causative agent of malaria, rely on glucose for energy supply during blood stage. Inhibition of glucose uptake thus represents a potential strategy for the development of antimalarial drugs. Here, we present the crystal structures of PfHT1, the sole hexose transporter in the genome of Plasmodium species, at resolutions of 2.6 Å in complex with D-glucose and 3.7 Å with a moderately selective inhibitor, C3361. Although both structures exhibit occluded conformations, binding of C3361 induces marked rearrangements that result in an additional pocket. This inhibitor-binding-induced pocket presents an opportunity for the rational design of PfHT1-specific inhibitors. Among our designed C3361 derivatives, several exhibited improved inhibition of PfHT1 and cellular potency against P. falciparum, with excellent selectivity to human GLUT1. These findings serve as a proof of concept for the development of the next-generation antimalarial chemotherapeutics by simultaneously targeting the orthosteric and allosteric sites of PfHT1.
- State Key Laboratory of Membrane Biology, Tsinghua University, Beijing 100084, China; Beijing Advanced Innovation Center for Structural Biology, Tsinghua University, Beijing 100084, China; Tsinghua-Peking Joint Center for Life Sciences, Tsinghua University, Beijing 100084, China; School of Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: