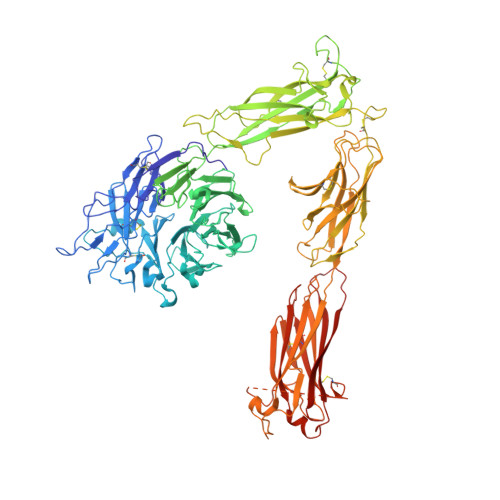
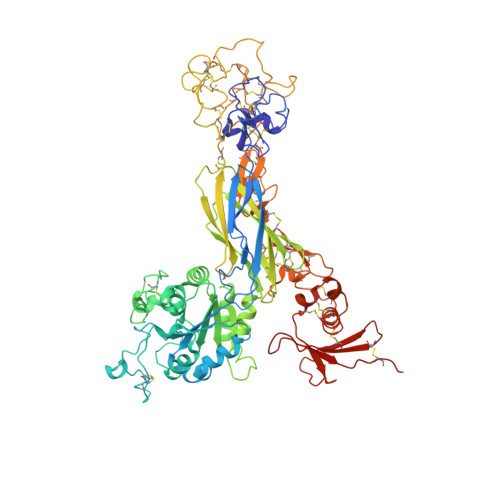
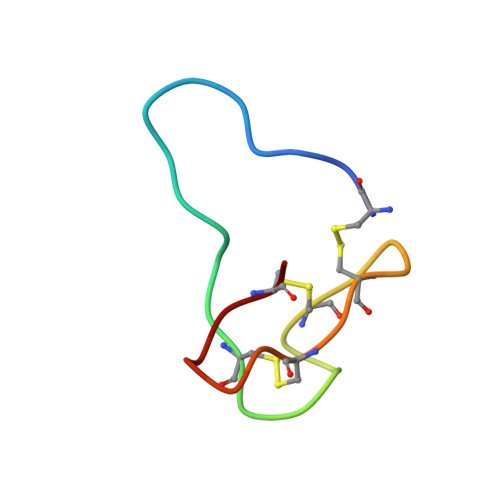
Structural Basis of the Differential Binding of Engineered Knottins to Integrins alpha V beta 3 and alpha 5 beta 1.
Van Agthoven, J.F., Shams, H., Cochran, F.V., Alonso, J.L., Kintzing, J.R., Garakani, K., Adair, B.D., Xiong, J.P., Mofrad, M.R.K., Cochran, J.R., Arnaout, M.A.(2019) Structure 27: 1443-1451.e6
- PubMed: 31353240
- DOI: https://doi.org/10.1016/j.str.2019.06.011
- Primary Citation of Related Structures:
6MSL, 6MSU - PubMed Abstract:
Targeting both integrins αVβ3 and α5β1 simultaneously appears to be more effective in cancer therapy than targeting each one alone. The structural requirements for bispecific binding of ligand to integrins have not been fully elucidated. RGD-containing knottin 2.5F binds selectively to αVβ3 and α5β1, whereas knottin 2.5D is αVβ3 specific. To elucidate the structural basis of this selectivity, we determined the structures of 2.5F and 2.5D as apo proteins and in complex with αVβ3, and compared their interactions with integrins using molecular dynamics simulations. These studies show that 2.5D engages αVβ3 by an induced fit, but conformational selection of a flexible RGD loop accounts for high-affinity selective binding of 2.5F to both integrins. The contrasting binding of the highly flexible low-affinity linear RGD peptides to multiple integrins suggests that a "Goldilocks zone" of conformational flexibility of the RGD loop in 2.5F underlies its selective binding promiscuity to integrins.
- Leukocyte Biology and Inflammation Program, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA 02129, USA; Structural Biology Program, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA 02129, USA; Division of Nephrology/Department of Medicine, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA 02129, USA.
Organizational Affiliation: