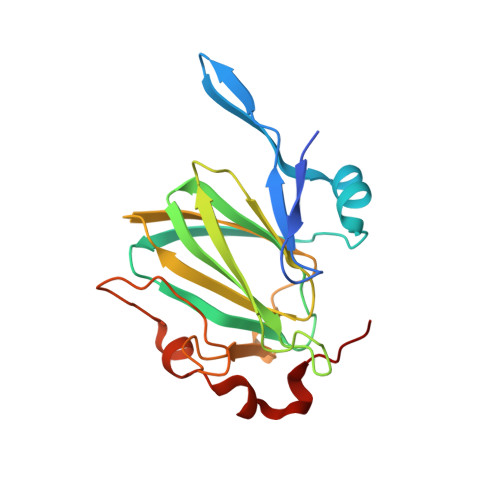
Crystal structure of dTDP-6-deoxy-D-glucose-3,5-epimerase RmlC from Legionella pneumophila Philadelphia 1 in complex with dTDP-4-KETO-L-RHAMNOSE
Abendroth, J., Schulze, M.-S., Mayclin, S.J., Lorimer, D.D., Horanyi, P.S., Edwards, T.E.To be published.