Mapping of a Novel H3-Specific Broadly Neutralizing Monoclonal Antibody Targeting the Hemagglutinin Globular Head Isolated from an Elite Influenza Virus-Immunized Donor Exhibiting Serological Breadth.
Qiu, Y., Stegalkina, S., Zhang, J., Boudanova, E., Park, A., Zhou, Y., Prabakaran, P., Pougatcheva, S., Ustyugova, I.V., Vogel, T.U., Mundle, S.T., Oomen, R., Delagrave, S., Ross, T.M., Kleanthous, H., Qiu, H.(2020) J Virol 94
- PubMed: 31826999
- DOI: https://doi.org/10.1128/JVI.01035-19
- Primary Citation of Related Structures:
6PDX - PubMed Abstract:
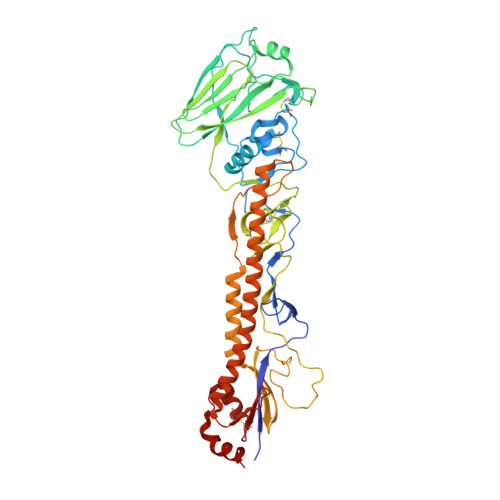
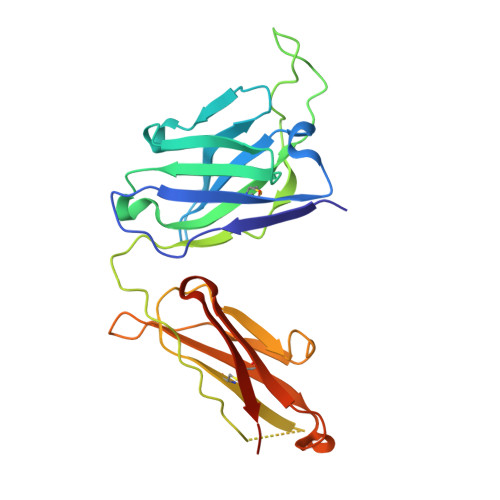
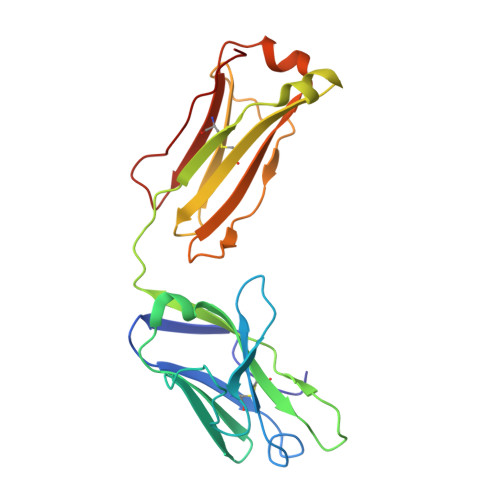
The discovery of potent and broadly protective influenza virus epitopes could lead to improved vaccines that are resistant to antigenic drift. Here, we describe human antibody C585, isolated from a vaccinee with remarkable serological breadth as measured by hemagglutinin inhibition (HAI). C585 binds and neutralizes multiple H3N2 strains isolated between 1968 and 2016, including strains that emerged up to 4 years after B cells were isolated from the vaccinated donor. The crystal structure of C585 Fab in complex with the HA from A/Switzerland/9715293/2013 (H3N2) shows that the antibody binds to a novel and well-conserved epitope on the globular head of H3 HA and that it differs from other antibodies not only in its epitope but in its binding geometry and hypermutated framework 3 region, thereby explaining its breadth and ability to mediate hemagglutination inhibition across decades of H3N2 strains. The existence of epitopes such as the one elucidated by C585 has implications for rational vaccine design. IMPORTANCE Influenza viruses escape immunity through continuous antigenic changes that occur predominantly on the viral hemagglutinin (HA). Induction of broadly neutralizing antibodies (bnAbs) targeting conserved epitopes following vaccination is a goal of universal influenza vaccines and advantageous in protecting hosts against virus evolution and antigenic drift. To date, most of the discovered bnAbs bind either to conserved sites in the stem region or to the sialic acid-binding pocket. Generally, antibodies targeting the stem region offer broader breadth with low potency, while antibodies targeting the sialic acid-binding pocket cover narrower breadth but usually have higher potency. In this study, we identified a novel neutralizing epitope in the head region recognized by a broadly neutralizing human antibody against a broad range of H3N2 with high potency. This epitope may provide insights for future universal vaccine design.
- Biologics Research, Sanofi, Framingham, Massachusetts, USA Yu.Qiu@sanofi.com harry.kleanthous@gatesfoundation.org.
Organizational Affiliation: