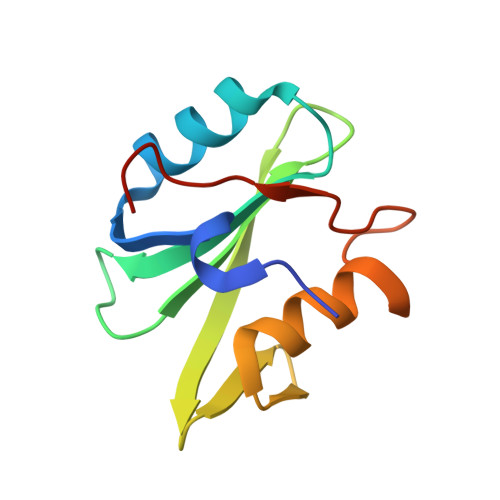
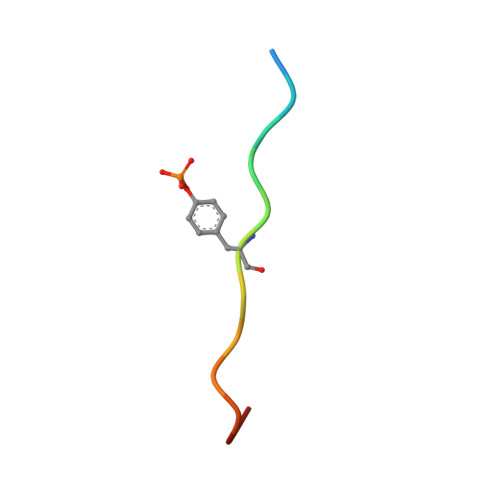
Crystal structures of p120RasGAP N-terminal SH2 domain in its apo form and in complex with a p190RhoGAP phosphotyrosine peptide.
Jaber Chehayeb, R., Stiegler, A.L., Boggon, T.J.(2019) PLoS One 14: e0226113-e0226113
- PubMed: 31891593
- DOI: https://doi.org/10.1371/journal.pone.0226113
- Primary Citation of Related Structures:
6PXB, 6PXC - PubMed Abstract:
The Rho and Ras pathways play vital roles in cell growth, division and motility. Cross-talk between the pathways amplifies their roles in cell proliferation and motility and its dysregulation is involved in disease pathogenesis. One important interaction for cross-talk occurs between p120RasGAP (RASA1), a GTPase activating protein (GAP) for Ras, and p190RhoGAP (p190RhoGAP-A, ARHGAP35), a GAP for Rho. The binding of these proteins is primarily mediated by two SH2 domains within p120RasGAP engaging phosphorylated tyrosines of p190RhoGAP, of which the best studied is pTyr-1105. To better understand the interaction between p120RasGAP and p190RhoGAP, we determined the 1.75 Å X-ray crystal structure of the N-terminal SH2 domain of p120RasGAP in the unliganded form, and its 1.6 Å co-crystal structure in complex with a synthesized phosphotyrosine peptide, EEENI(p-Tyr)SVPHDST, corresponding to residues 1100-1112 of p190RhoGAP. We find that the N-terminal SH2 domain of p120RhoGAP has the characteristic SH2 fold encompassing a central beta-sheet flanked by two alpha-helices, and that peptide binding stabilizes specific conformations of the βE-βF loop and arginine residues R212 and R231. Site-directed mutagenesis and native gel shifts confirm phosphotyrosine binding through the conserved FLVR motif arginine residue R207, and isothermal titration calorimetry finds a dissociation constant of 0.3 ± 0.1 μM between the phosphopeptide and SH2 domain. These results demonstrate that the major interaction between two important GAP proteins, p120RasGAP and p190RhoGAP, is mediated by a canonical SH2-pTyr interaction.
- Yale College, New Haven, Connecticut, United States of America.
Organizational Affiliation: