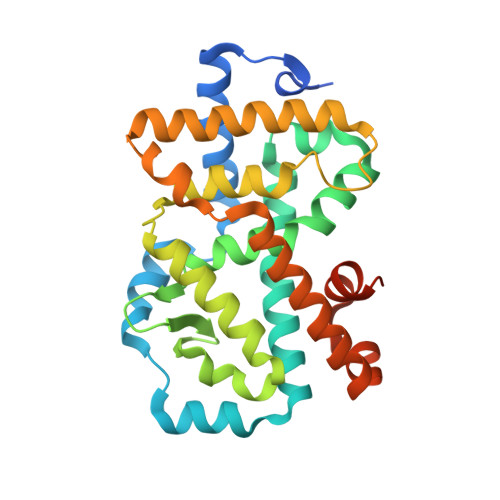
Discovery of Potent and Orally Bioavailable Inverse Agonists of the Retinoic Acid Receptor-Related Orphan Receptor C2.
von Berg, S., Xue, Y., Collins, M., Llinas, A., Olsson, R.I., Halvarsson, T., Lindskog, M., Malmberg, J., Jirholt, J., Krutrok, N., Ramnegard, M., Brannstrom, M., Lundqvist, A., Lepisto, M., Aagaard, A., McPheat, J., Hansson, E.L., Chen, R., Xiong, Y., Hansson, T.G., Narjes, F.(2019) ACS Med Chem Lett 10: 972-977
- PubMed: 31223457
- DOI: https://doi.org/10.1021/acsmedchemlett.9b00158
- Primary Citation of Related Structures:
6R7A, 6R7J, 6R7K - PubMed Abstract:
The further optimization of a recently disclosed series of inverse agonists of the nuclear receptor RORC2 is described. Investigations into the left-hand side of compound 1 , guided by X-ray crystal structures, led to the substitution of the 4-aryl-thiophenyl residue with the hexafluoro-2-phenyl-propan-2-ol moiety. This change resulted in to compound 28 , which combined improved drug-like properties with good cell potency and a significantly lower dose, using an early dose to man prediction. Target engagement in vivo was demonstrated in the thymus of mice by a reduction in the number of double positive T cells after oral dosing.
- Medicinal Chemistry, DMPK, andBioscience, Research and Early Development, Respiratory, Inflammation and Autoimmune, BioPharmaceuticals R&D, AstraZeneca, Gothenburg, SE-43183 Mölndal, Sweden.
Organizational Affiliation: