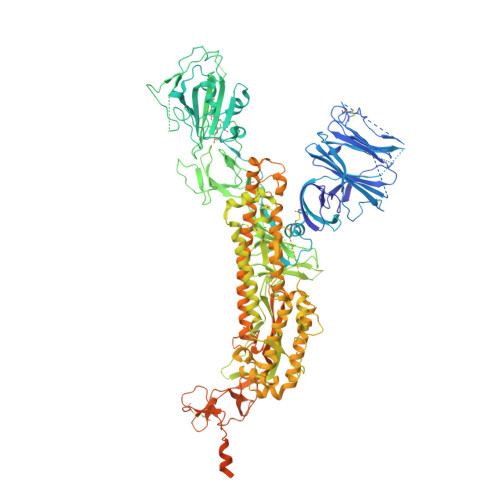
Controlling the SARS-CoV-2 spike glycoprotein conformation.
Henderson, R., Edwards, R.J., Mansouri, K., Janowska, K., Stalls, V., Gobeil, S.M.C., Kopp, M., Li, D., Parks, R., Hsu, A.L., Borgnia, M.J., Haynes, B.F., Acharya, P.(2020) Nat Struct Mol Biol 27: 925-933
- PubMed: 32699321
- DOI: https://doi.org/10.1038/s41594-020-0479-4
- Primary Citation of Related Structures:
6X29, 6X2A, 6X2B, 6X2C - PubMed Abstract:
The coronavirus (CoV) spike (S) protein, involved in viral-host cell fusion, is the primary immunogenic target for virus neutralization and the current focus of many vaccine design efforts. The highly flexible S-protein, with its mobile domains, presents a moving target to the immune system. Here, to better understand S-protein mobility, we implemented a structure-based vector analysis of available β-CoV S-protein structures. Despite an overall similarity in domain organization, we found that S-proteins from different β-CoVs display distinct configurations. Based on this analysis, we developed two soluble ectodomain constructs for the SARS-CoV-2 S-protein, in which the highly immunogenic and mobile receptor binding domain (RBD) is either locked in the all-RBDs 'down' position or adopts 'up' state conformations more readily than the wild-type S-protein. These results demonstrate that the conformation of the S-protein can be controlled via rational design and can provide a framework for the development of engineered CoV S-proteins for vaccine applications.
- Duke Human Vaccine Institute, Durham, NC, USA. rory.henderson@duke.edu.
Organizational Affiliation: