Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sacco, M.D., Ma, C., Lagarias, P., Gao, A., Townsend, J.A., Meng, X., Dube, P., Zhang, X., Hu, Y., Kitamura, N., Hurst, B., Tarbet, B., Marty, M.T., Kolocouris, A., Xiang, Y., Chen, Y., Wang, J.(2020) Sci Adv 6
- PubMed: 33158912
- DOI: https://doi.org/10.1126/sciadv.abe0751
- Primary Citation of Related Structures:
6XA4, 6XBG, 6XBH, 6XBI, 6XFN - PubMed Abstract:
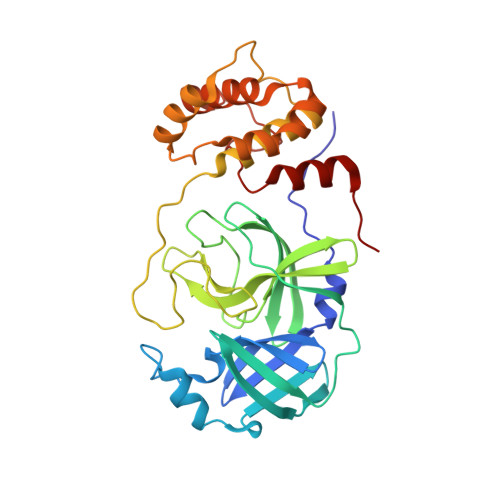
The main protease (M pro ) of SARS-CoV-2 is a key antiviral drug target. While most M pro inhibitors have a γ-lactam glutamine surrogate at the P1 position, we recently found that several M pro inhibitors have hydrophobic moieties at the P1 site, including calpain inhibitors II and XII, which are also active against human cathepsin L, a host protease that is important for viral entry. In this study, we solved x-ray crystal structures of M pro in complex with calpain inhibitors II and XII and three analogs of GC-376 The structure of M pro with calpain inhibitor II confirmed that the S1 pocket can accommodate a hydrophobic methionine side chain, challenging the idea that a hydrophilic residue is necessary at this position. The structure of calpain inhibitor XII revealed an unexpected, inverted binding pose. Together, the biochemical, computational, structural, and cellular data presented herein provide new directions for the development of dual inhibitors as SARS-CoV-2 antivirals.
- Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA.
Organizational Affiliation: