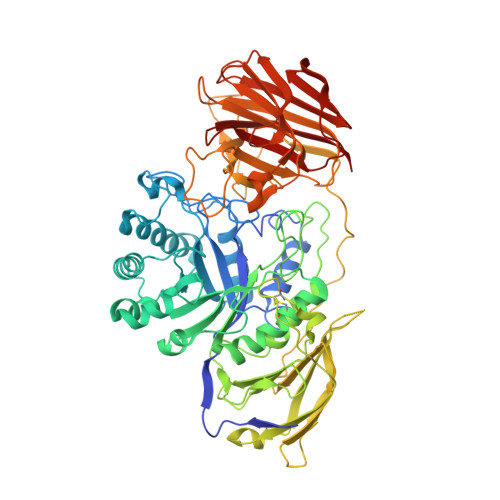
The Bicyclic Form of galacto -Noeurostegine Is a Potent Inhibitor of beta-Galactocerebrosidase.
Viuff, A., Salamone, S., McLoughlin, J., Deane, J.E., Jensen, H.H.(2021) ACS Med Chem Lett 12: 56-59
- PubMed: 33488964
- DOI: https://doi.org/10.1021/acsmedchemlett.0c00377
- Primary Citation of Related Structures:
6Y6S, 6Y6T - PubMed Abstract:
Competitive inhibitors of galactocerebrosidase (GALC) could be candidates for pharmacological chaperone therapy of patients with Krabbe disease. The known and selective nortropane-type iminosugar galacto -noeurostegine has been found to competitively inhibit GALC with K i = 7 μM at pH 4.6, which is 330-fold more potent than the analogous deoxynoeurostegine. It was shown through X-ray protein crystallography that galacto -noeurostegine binds to the active site of GALC in its bicyclic form.
Organizational Affiliation:
Department of Chemistry, Aarhus University, Langelandsgade 140, 8000 Aarhus C, Denmark.