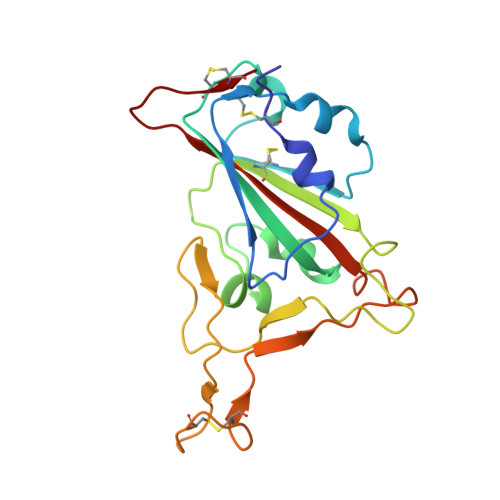
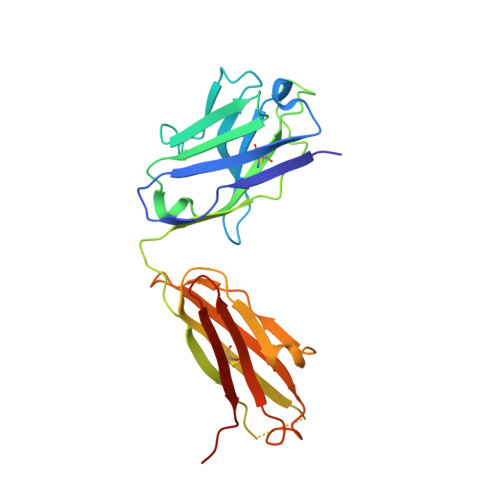
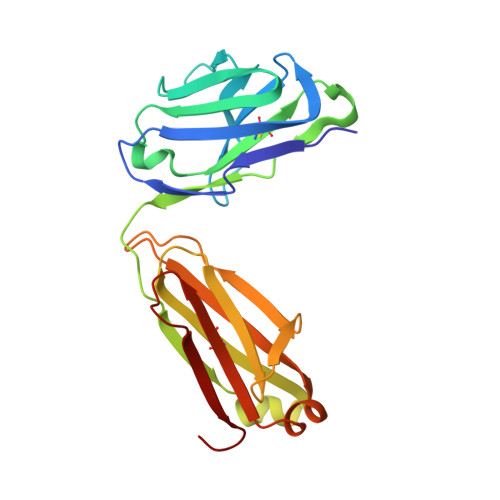
Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Zhou, D., Duyvesteyn, H.M.E., Chen, C.P., Huang, C.G., Chen, T.H., Shih, S.R., Lin, Y.C., Cheng, C.Y., Cheng, S.H., Huang, Y.C., Lin, T.Y., Ma, C., Huo, J., Carrique, L., Malinauskas, T., Ruza, R.R., Shah, P.N.M., Tan, T.K., Rijal, P., Donat, R.F., Godwin, K., Buttigieg, K.R., Tree, J.A., Radecke, J., Paterson, N.G., Supasa, P., Mongkolsapaya, J., Screaton, G.R., Carroll, M.W., Gilbert-Jaramillo, J., Knight, M.L., James, W., Owens, R.J., Naismith, J.H., Townsend, A.R., Fry, E.E., Zhao, Y., Ren, J., Stuart, D.I., Huang, K.A.(2020) Nat Struct Mol Biol 27: 950-958
- PubMed: 32737466
- DOI: https://doi.org/10.1038/s41594-020-0480-y
- Primary Citation of Related Structures:
6ZCZ, 6ZDG, 6ZDH, 6ZER, 6ZFO - PubMed Abstract:
The COVID-19 pandemic has had an unprecedented health and economic impact and there are currently no approved therapies. We have isolated an antibody, EY6A, from an individual convalescing from COVID-19 and have shown that it neutralizes SARS-CoV-2 and cross-reacts with SARS-CoV-1. EY6A Fab binds the receptor binding domain (RBD) of the viral spike glycoprotein tightly (K D of 2 nM), and a 2.6-Å-resolution crystal structure of an RBD-EY6A Fab complex identifies the highly conserved epitope, away from the ACE2 receptor binding site. Residues within this footprint are key to stabilizing the pre-fusion spike. Cryo-EM analyses of the pre-fusion spike incubated with EY6A Fab reveal a complex of the intact spike trimer with three Fabs bound and two further multimeric forms comprising the destabilized spike attached to Fab. EY6A binds what is probably a major neutralizing epitope, making it a candidate therapeutic for COVID-19.
- Division of Structural Biology, Nuffield Department of Medicine, University of Oxford, The Wellcome Centre for Human Genetics, Headington, Oxford, UK.
Organizational Affiliation: