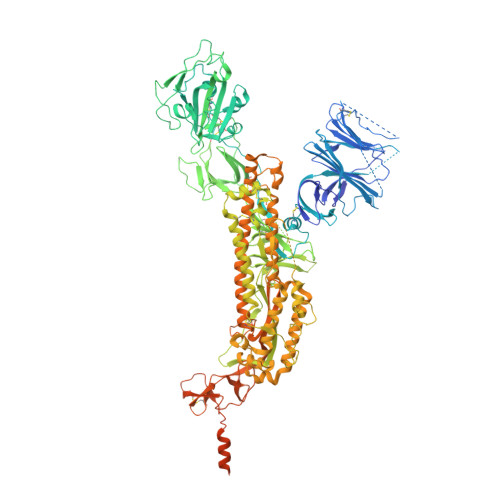
Structures and distributions of SARS-CoV-2 spike proteins on intact virions.
Ke, Z., Oton, J., Qu, K., Cortese, M., Zila, V., McKeane, L., Nakane, T., Zivanov, J., Neufeldt, C.J., Cerikan, B., Lu, J.M., Peukes, J., Xiong, X., Krausslich, H.G., Scheres, S.H.W., Bartenschlager, R., Briggs, J.A.G.(2020) Nature 588: 498-502
- PubMed: 32805734
- DOI: https://doi.org/10.1038/s41586-020-2665-2
- Primary Citation of Related Structures:
6ZWV - PubMed Abstract:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virions are surrounded by a lipid bilayer from which spike (S) protein trimers protrude 1 . Heavily glycosylated S trimers bind to the angiotensin-converting enzyme 2 receptor and mediate entry of virions into target cells 2-6 . S exhibits extensive conformational flexibility: it modulates exposure of its receptor-binding site and subsequently undergoes complete structural rearrangement to drive fusion of viral and cellular membranes 2,7,8 . The structures and conformations of soluble, overexpressed, purified S proteins have been studied in detail using cryo-electron microscopy 2,7,9-12 , but the structure and distribution of S on the virion surface remain unknown. Here we applied cryo-electron microscopy and tomography to image intact SARS-CoV-2 virions and determine the high-resolution structure, conformational flexibility and distribution of S trimers in situ on the virion surface. These results reveal the conformations of S on the virion, and provide a basis from which to understand interactions between S and neutralizing antibodies during infection or vaccination.
- Structural Studies Division, Medical Research Council Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: