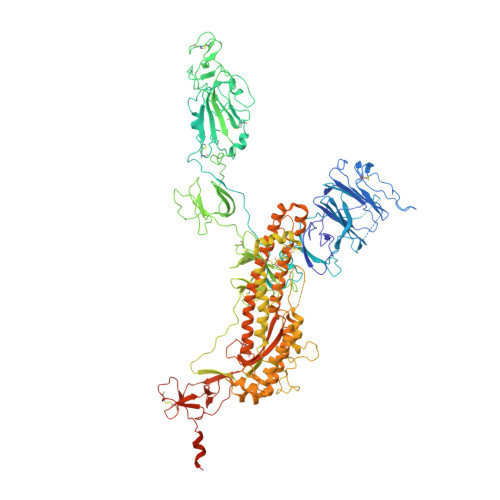
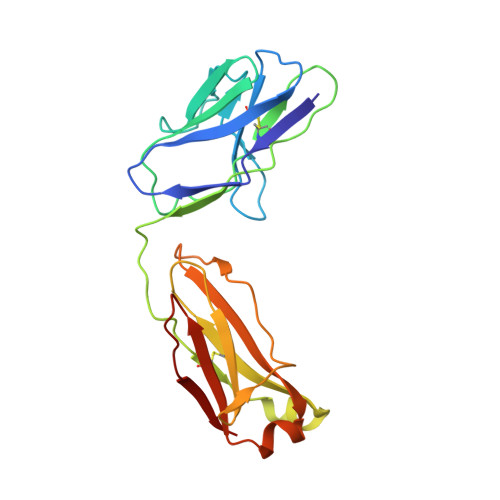
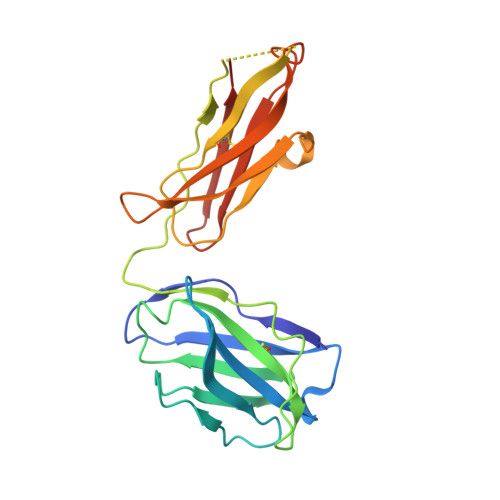
Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Lv, Z., Deng, Y.Q., Ye, Q., Cao, L., Sun, C.Y., Fan, C., Huang, W., Sun, S., Sun, Y., Zhu, L., Chen, Q., Wang, N., Nie, J., Cui, Z., Zhu, D., Shaw, N., Li, X.F., Li, Q., Xie, L., Wang, Y., Rao, Z., Qin, C.F., Wang, X.(2020) Science 369: 1505-1509
- PubMed: 32703908
- DOI: https://doi.org/10.1126/science.abc5881
- Primary Citation of Related Structures:
7CAB, 7CAC, 7CAH, 7CAI, 7CAK - PubMed Abstract:
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in an unprecedented public health crisis. There are no approved vaccines or therapeutics for treating COVID-19. Here we report a humanized monoclonal antibody, H014, that efficiently neutralizes SARS-CoV-2 and SARS-CoV pseudoviruses as well as authentic SARS-CoV-2 at nanomolar concentrations by engaging the spike (S) receptor binding domain (RBD). H014 administration reduced SARS-CoV-2 titers in infected lungs and prevented pulmonary pathology in a human angiotensin-converting enzyme 2 mouse model. Cryo-electron microscopy characterization of the SARS-CoV-2 S trimer in complex with the H014 Fab fragment unveiled a previously uncharacterized conformational epitope, which was only accessible when the RBD was in an open conformation. Biochemical, cellular, virological, and structural studies demonstrated that H014 prevents attachment of SARS-CoV-2 to its host cell receptors. Epitope analysis of available neutralizing antibodies against SARS-CoV and SARS-CoV-2 uncovered broad cross-protective epitopes. Our results highlight a key role for antibody-based therapeutic interventions in the treatment of COVID-19.
- CAS Key Laboratory of Infection and Immunity, National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: