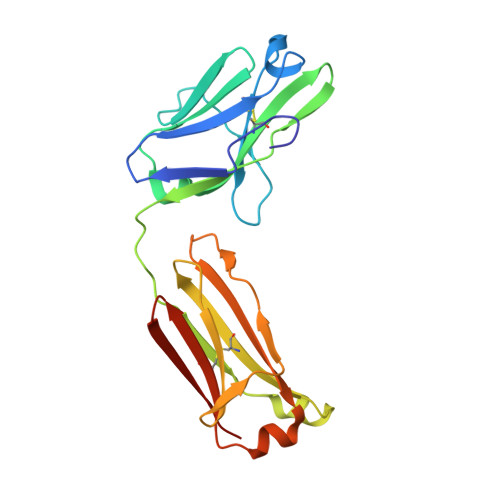
Crystal structure of human immunoglobulin fragment Fab New refined at 2.0 A resolution.
Saul, F.A., Poljak, R.J.(1992) Proteins 14: 363-371
- PubMed: 1438175
- DOI: https://doi.org/10.1002/prot.340140305
- Primary Citation of Related Structures:
7FAB - PubMed Abstract:
The three-dimensional structure of the human immunoglobulin fragment Fab New (IgG1, lambda) has been refined to a crystallographic R-factor of 16.9% to 2 A resolution. Rms deviations of the final model from ideal geometry are 0.014 A for bond distances and 3.03 degrees for bond angles. Refinement was based on a new X-ray data set including 28,301 reflections with F > 2.5 sigma(F) from 6.0 to 2.0 A resolution. The starting model for the refinement procedure reported here is from the Brookhaven Protein Data Bank entry 3FAB (rev. 1981). Differences between the initial and final models include modified polypeptide-chain folding in the third complementarity-determining region (CDR3) and the third framework region (FR3) of VH and in some exposed loops of CL and CH1. Amino acid sequence changes were determined at a number of positions by inspection of difference electron density maps. The incorporation of amino acid sequence changes results in an improved VH framework model for the "humanization" of monoclonal antibodies.
- Département d'Immunologie, Institut Pasteur, Paris, France.
Organizational Affiliation: