High-resolution three-dimensional structure of interleukin 1 beta in solution by three- and four-dimensional nuclear magnetic resonance spectroscopy.
Clore, G.M., Wingfield, P.T., Gronenborn, A.M.(1991) Biochemistry 30: 2315-2323
- PubMed: 2001363
- DOI: https://doi.org/10.1021/bi00223a005
- Primary Citation of Related Structures:
6I1B, 7I1B - PubMed Abstract:
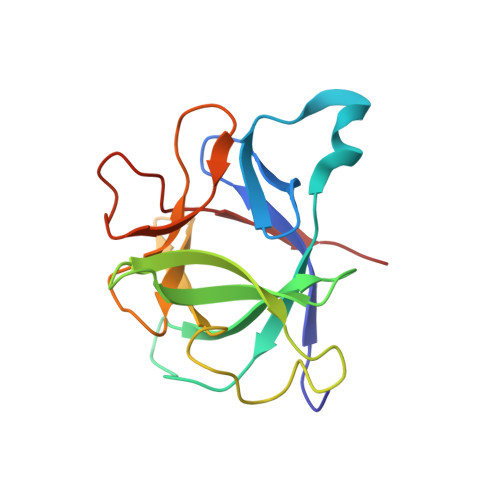
The determination of the high-resolution three-dimensional solution structure of interleukin 1 beta (IL-1 beta), a protein of 153 residues and 17.4 kDa, which plays a central role in the immune and inflammatory responses, has been determined by heteronuclear (13C and 15N) three- and four-dimensional NMR spectroscopy. The structure is based on 3146 experimental restraints comprising 2780 distance and 366 torsion angle (phi, psi, and chi 1) restraints. A total of 32 simulated annealing structures are calculated, and the atomic RMS distribution about the mean coordinate positions is 0.41 +/- 0.04 A for the backbone atoms and 0.82 +/- 0.04 A for all atoms (excluding residue 1 at the N-terminus and residues 152 and 153 at the C-terminus, which are partially disordered). In the case of internal side chains with a surface accessibility of less than or equal to 40%, the atomic RMS distribution about the mean coordinate positions for all atoms is 0.49 +/- 0.03 A. IL-1 beta resembles a tetrahedron and is composed of 12 beta-strands arranged in three pseudosymmetric topological units, each of which comprises 5 strands. Analysis of the mutational data on IL-1 beta in the light of the three-dimensional structure suggests the presence of three distinct binding sites for the IL-1 receptor on the surface of the protein. It is suggested that each of the three immunoglobulin domains which comprise the extracellular portion of the IL-1 receptor recognizes one of these sites.
- Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892.
Organizational Affiliation: