Crystal structure of Paradendryphiella salina PL7A alginate lyase mutant Y223F in complex with tri-mannuronic acid
Fredslund, F., Welner, D.H., Wilkens, C.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Alginate lyase (PL7) | 231 | Paradendryphiella salina | Mutation(s): 1 Gene Names: PsAlg7A EC: 4.2.2.3 | 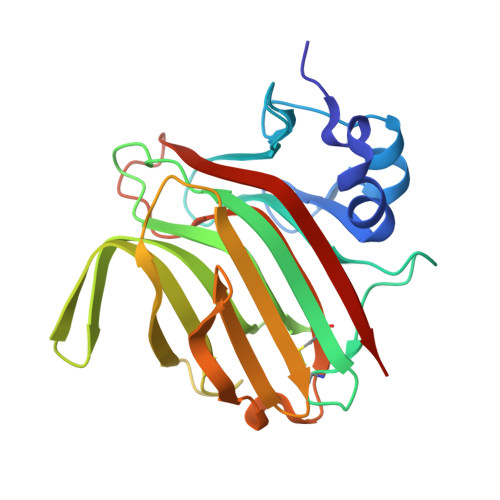 | |
UniProt | |||||
Find proteins for A0A485PVH1 (Paradendryphiella salina) Explore A0A485PVH1 Go to UniProtKB: A0A485PVH1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A485PVH1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MAV (Subject of Investigation/LOI) Query on MAV | C [auth A] | alpha-D-mannopyranuronic acid C6 H10 O7 AEMOLEFTQBMNLQ-BYHBOUFCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 34.98 | α = 90 |
| b = 81.22 | β = 90 |
| c = 80.8 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| MxCuBE | data collection |
| XDS | data scaling |
| Coot | model building |
| PHASER | phasing |
| XDS | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Union (EU) | Denmark | 9082-00021B |