A Multi-Specific DARPin Potently Neutralizes Shiga Toxin 2 via Simultaneous Modulation of Both Toxin Subunits.
Zeng, Y., Jiang, M., Robinson, S., Peng, Z., Chonira, V., Simeon, R., Tzipori, S., Zhang, J., Chen, Z.(2022) Bioengineering (Basel) 9
- PubMed: 36290479
- DOI: https://doi.org/10.3390/bioengineering9100511
- Primary Citation of Related Structures:
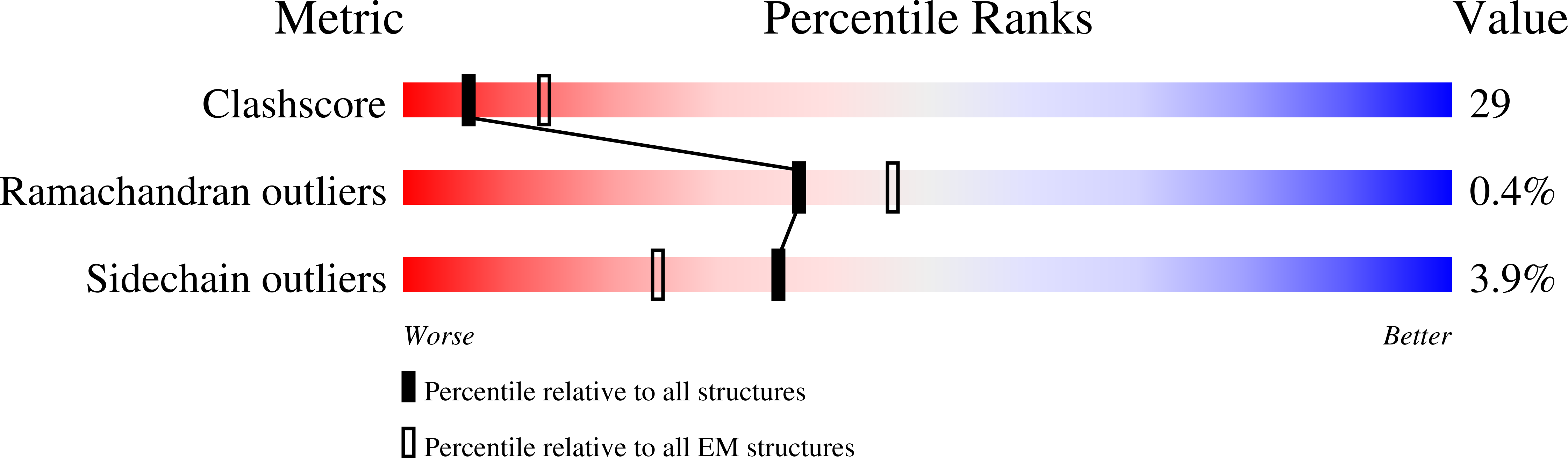
7UJJ - PubMed Abstract:
Shiga toxin-producing E. coli (STEC) is a common cause of bloody diarrhea. The pathology of STEC infection derives from two exotoxins-Shiga toxin 1 (Stx1) and Shiga toxin 2 (Stx2)-that are secreted by STEC in the gut, from where they are systemically absorbed, causing severe kidney damage leading to hemolytic uremic syndrome (HUS). Currently, there is no effective treatment for HUS, and only supportive care is recommended. We report the engineering of a panel of designed ankyrin repeat proteins (DARPin) with potent neutralization activity against Stx2a, the major subtype associated with HUS. The best dimeric DARPin, SD5, created via a combination of directed evolution and rational design, neutralizes Stx2a with a half maximal effective concentration (EC 50 ) of 0.61 nM in vitro. The two monomeric DARPin constituents of SD5 exhibit complementary functions-SHT targets the enzymatic A subunit of Stx2a and inhibits the toxin's catalytic activity, while DARPin #3 binds the B subunit, based on the cryo-EM study, and induces a novel conformational change in the B subunit that distorts its five-fold symmetry and presumably interferes with toxin attachment to target cells. SD5 was fused to an albumin-binding DARPin, and the resulting trimeric DARPin DA1-SD5 efficiently protects mice in a toxin challenge model, pointing to a high potential of this DARPin as a therapeutic for STEC infection. Finally, the unprecedented toxin conformational change induced by DARPin #3 represents a novel mode of action for neutralizing Stx2 toxicity and reveals new targets for future drug development.
Organizational Affiliation:
Department of Microbial Pathogenesis and Immunology, Texas A&M University Health Science Center, 8847 Riverside Pkwy, Bryan, TX 77807, USA.