An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Ge, Q., Teng, M., Li, X., Guo, Q., Tao, Y.(2023) Structure 31: 33-43.e5
- PubMed: 36513066
- DOI: https://doi.org/10.1016/j.str.2022.11.009
- Primary Citation of Related Structures:
7X8P, 7X8Q, 7X8T - PubMed Abstract:
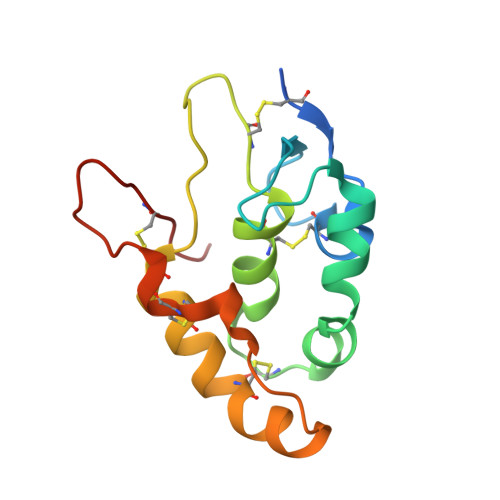
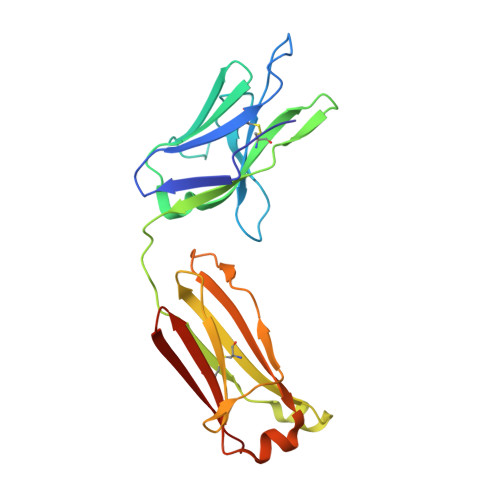
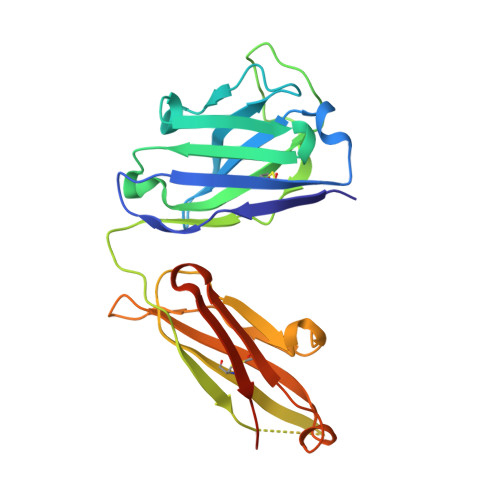
The lack of incorporating epitope information into the selection process makes the conventional antibody screening method less effective in identifying antibodies with desired functions. Here, we developed an epitope-directed antibody selection method by designing a directed library favoring the target epitope and a precise "counter" antigen for clearing irrelevant binders in the library. With this method, we successfully isolated an antibody, pF7_A5, that targets the less conserved region on the FZD2/7 CRD as designed. Guided by the structure of pF7_A5-FZD2 CRD , a further round of evolution was conducted together with the "counter" antigen selection strategy, and ultimately, an FZD2-specific antibody and an FZD7-preferred antibody were obtained. Because of targeting the predefined functional site, all these antibodies exhibited the expected modulatory activity on the Wnt pathway. Together, the method developed here will be useful in antibody drug discovery, and the identified FZD antibodies will have clinical potential in FZD-related cancer therapy.
Organizational Affiliation:
Department of Clinical Laboratory, The First Affiliated Hospital of USTC, Ministry of Education Key Laboratory for Membraneless Organelles & Cellular Dynamics, Biomedical Sciences and Health Laboratory of Anhui Province, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, 230027 Hefei, P.R. China.