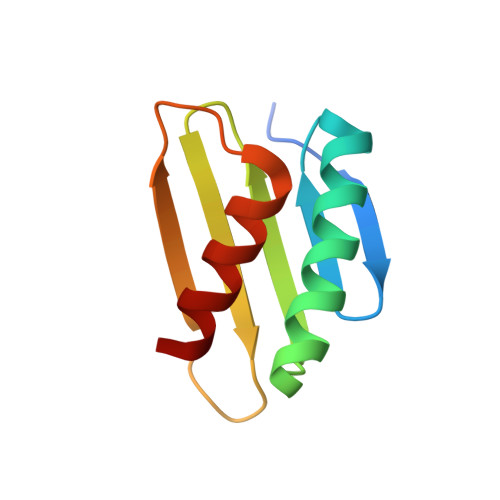
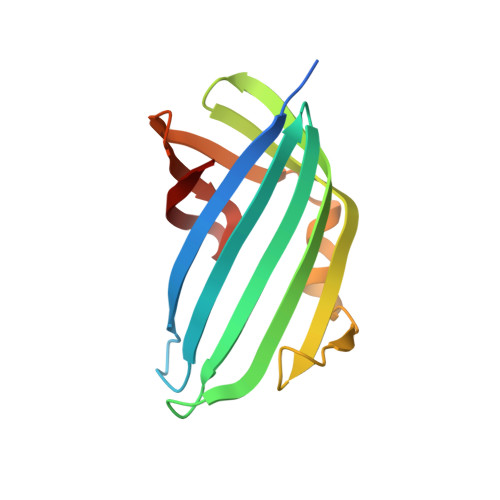
Molecular determinants for differential activation of the bile acid receptor from the pathogen Vibrio parahaemolyticus.
Zou, A.J., Kinch, L., Chimalapati, S., Garcia, N., Tomchick, D.R., Orth, K.(2023) J Biol Chem 299: 104591-104591
- PubMed: 36894018
- DOI: https://doi.org/10.1016/j.jbc.2023.104591
- Primary Citation of Related Structures:
8DML - PubMed Abstract:
Bile acids are important for digestion of food and antimicrobial activity. Pathogenic Vibrio parahaemolyticus senses bile acids and induce pathogenesis. The bile acid taurodeoxycholate (TDC) was shown to activate the master regulator, VtrB, of this system, whereas other bile acids such as chenodeoxycholate (CDC) do not. Previously, VtrA-VtrC was discovered to be the co-component signal transduction system that binds bile acids and induces pathogenesis. TDC binds to the periplasmic domain of the VtrA-VtrC complex, activating a DNA-binding domain in VtrA that then activates VtrB. Here, we find that CDC and TDC compete for binding to the VtrA-VtrC periplasmic heterodimer. Our crystal structure of the VtrA-VtrC heterodimer bound to CDC revealed CDC binds in the same hydrophobic pocket as TDC but differently. Using isothermal titration calorimetry, we observed that most mutants in the binding pocket of VtrA-VtrC caused a decrease in bile acid binding affinity. Notably, two mutants in VtrC bound bile acids with a similar affinity as the WT protein but were attenuated for TDC-induced type III secretion system 2 activation. Collectively, these studies provide a molecular explanation for the selective pathogenic signaling by V. parahaemolyticus and reveal insight into a host's susceptibility to disease.
Organizational Affiliation:
Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, Texas, USA.