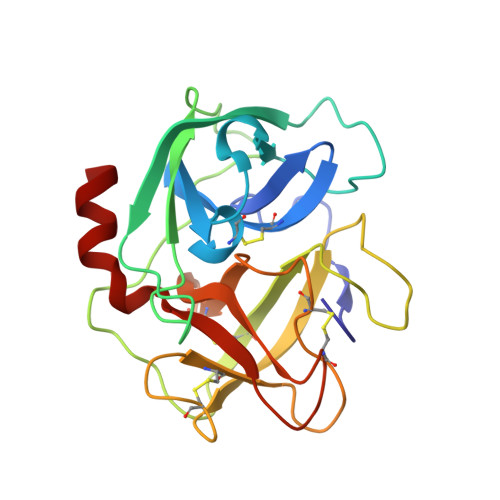
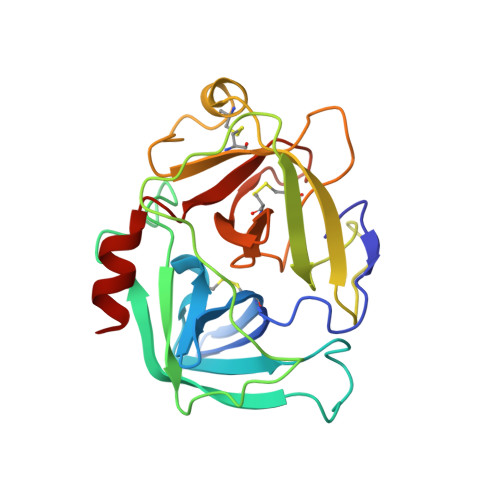
Simultaneous inhibition of two neutrophil serine proteases by the S. aureus innate immune evasion protein EapH2.
Mishra, N., Herdendorf, T.J., Prakash, O., Geisbrecht, B.V.(2023) J Biological Chem 299: 104878-104878
- PubMed: 37269950
- DOI: https://doi.org/10.1016/j.jbc.2023.104878
- Primary Citation of Related Structures:
8G24, 8G25, 8G26, 8GDG, 8GDH - PubMed Abstract:
Extracellular adherence protein domain (EAP) proteins are high-affinity, selective inhibitors of neutrophil serine proteases (NSP), including cathepsin-G (CG) and neutrophil elastase (NE). Most Staphylococcus aureus isolates encode for two EAPs, EapH1 and EapH2, that contain a single functional domain and share 43% identity with one another. Although structure/function investigations from our group have shown that EapH1 uses a globally similar binding mode to inhibit CG and NE, NSP inhibition by EapH2 is incompletely understood due to a lack of NSP/EapH2 cocrystal structures. To address this limitation, we further studied NSP inhibition by EapH2 in comparison with EapH1. Like its effects on NE, we found that EapH2 is a reversible, time-dependent, and low nanomolar affinity inhibitor of CG. We characterized an EapH2 mutant which suggested that the CG binding mode of EapH2 is comparable to EapH1. To test this directly, we used NMR chemical shift perturbation to study EapH1 and EapH2 binding to CG and NE in solution. Although we found that overlapping regions of EapH1 and EapH2 were involved in CG binding, we found that altogether distinct regions of EapH1 and EapH2 experienced changes upon binding to NE. An important implication of this observation is that EapH2 might be capable of binding and inhibiting CG and NE simultaneously. We confirmed this unexpected feature by solving crystal structures of the CG/EapH2/NE complex and demonstrating their functional relevance through enzyme inhibition assays. Together, our work defines a new mechanism of simultaneous inhibition of two serine proteases by a single EAP protein.
- Department of Biochemistry & Molecular Biophysics, Kansas State University, Manhattan, Kansas, USA.
Organizational Affiliation: