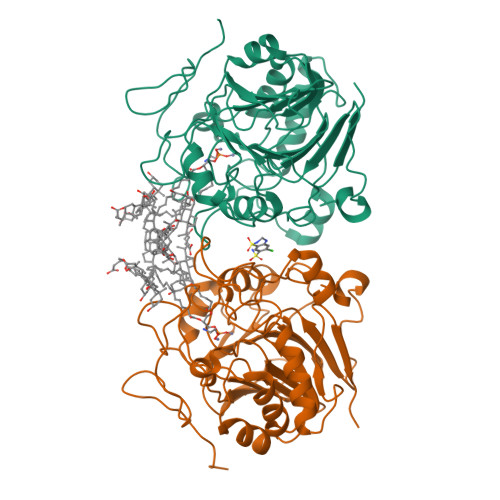
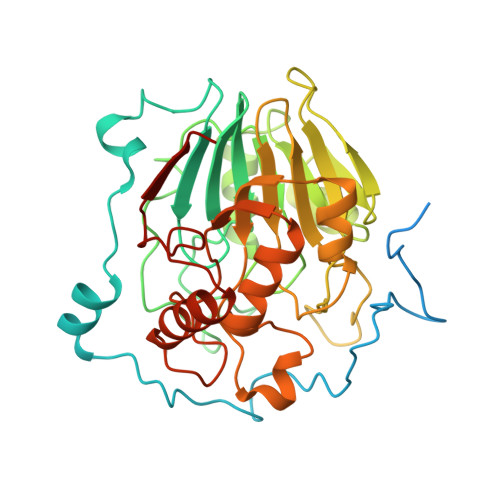
NAPE-PLD is target of thiazide diuretics
Chiarugi, S., Margheriti, F., De Lorenzi, V., Martino, E., Margheritis, E.G., Moscardini, A., Marotta, R., Chaves-Sanjuan, A., Del Seppia, C., Federighi, G., Lapi, D., Bandiera, T., Rapposelli, S., Scuri, R., Bolognesi, M., Garau, G.(2025) Cell Chem Biol