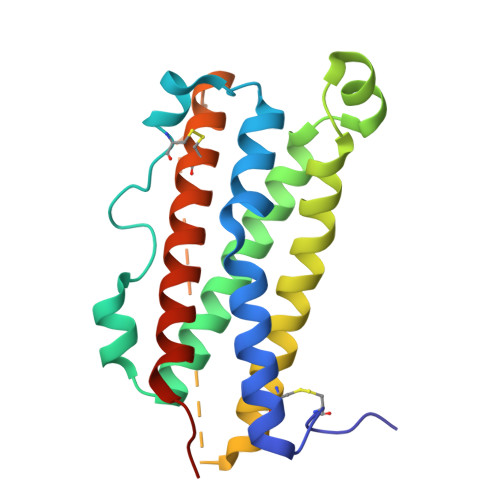
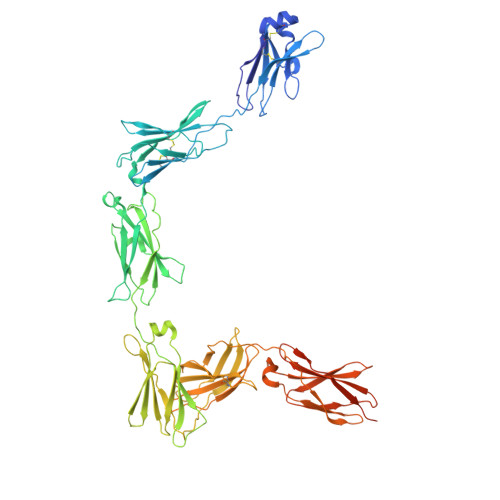
Structures of complete extracellular assemblies of type I and type II Oncostatin M receptor complexes.
Zhou, Y., Stevis, P.E., Cao, J., Ehrlich, G., Jones, J., Rafique, A., Sleeman, M.W., Olson, W.C., Franklin, M.C.(2024) Nat Commun 15: 9776-9776
- PubMed: 39532904
- DOI: https://doi.org/10.1038/s41467-024-54124-1
- Primary Citation of Related Structures:
8V29, 8V2A, 8V2B, 8V2C - PubMed Abstract:
Oncostatin M (OSM) is a unique Interleukin 6 (IL-6) family cytokine that plays pivotal roles in numerous biological events by signaling via two types of receptor complexes. While type I OSM receptor complex is formed by glycoprotein 130 (gp130) heterodimerization with Leukemia Inhibitory Factor receptor (LIFR), type II OSM receptor complex is composed of gp130 and OSM receptor (OSMR). OSM is an important contributor to multiple inflammatory diseases and cancers while OSM inhibition has been shown to be effective at reducing symptoms, making OSM an attractive therapeutic target. Using cryogenic electron microscopy (cryo-EM), we characterize full extracellular assemblies of human type I OSM receptor complex and mouse type II OSM receptor complex. The juxtamembrane domains of both complexes are situated in close proximity due to acute bends of the receptors. The rigid N-terminal extension of OSM contributes to gp130 binding and OSM signaling. Neither glycosylation nor pro-domain cleavage of OSM affects its activity. Mutagenesis identifies multiple OSM and OSMR residues crucial for complex formation and signaling. Our data reveal the structural basis for the assemblies of both type I and type II OSM receptor complexes and provide insights for modulation of OSM signaling in therapeutics.
- Regeneron Pharmaceuticals, Inc., Tarrytown, NY, 10591, USA. yi.zhou@regeneron.com.
Organizational Affiliation: