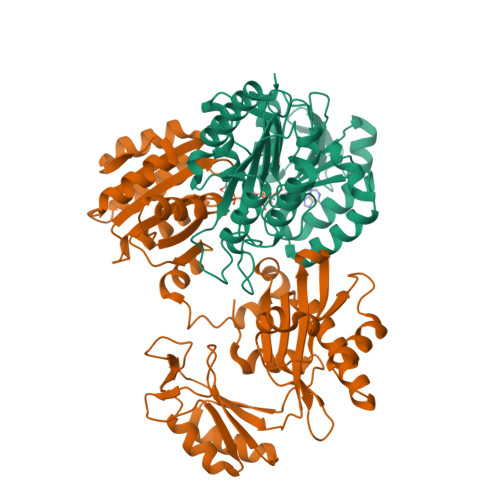
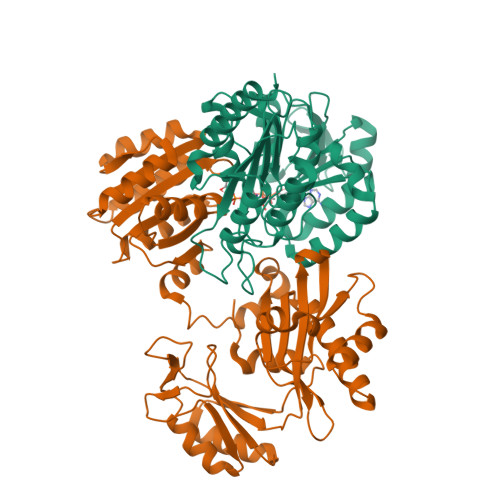
Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis.
Liu, R., Ren, X., Park, Y.E., Feng, H., Sheng, X., Song, X., AminiTabrizi, R., Shah, H., Li, L., Zhang, Y., Abdullah, K.G., Dubois-Coyne, S., Lin, H., Cole, P.A., DeBerardinis, R.J., McBrayer, S.K., Huang, H., Zhao, Y.(2025) Cell Metab 37: 377
- PubMed: 39642882
- DOI: https://doi.org/10.1016/j.cmet.2024.11.005
- Primary Citation of Related Structures:
8Z02, 8Z03 - PubMed Abstract:
Histone lysine lactylation is a physiologically and pathologically relevant epigenetic pathway that can be stimulated by the Warburg effect-associated L-lactate. Nevertheless, the mechanism by which cells use L-lactate to generate lactyl-coenzyme A (CoA) and how this process is regulated remains unknown. Here, we report the identification of guanosine triphosphate (GTP)-specific SCS (GTPSCS) as a lactyl-CoA synthetase in the nucleus. The mechanism was elucidated through the crystallographic structure of GTPSCS in complex with L-lactate, followed by mutagenesis experiments. GTPSCS translocates into the nucleus and interacts with p300 to elevate histone lactylation but not succinylation. This process depends on a nuclear localization signal in the GTPSCS G1 subunit and acetylation at G2 subunit residue K73, which mediates the interaction with p300. GTPSCS/p300 collaboration synergistically regulates histone H3K18la and GDF15 expression, promoting glioma proliferation and radioresistance. GTPSCS represents the inaugural enzyme to catalyze lactyl-CoA synthesis for epigenetic histone lactylation and regulate oncogenic gene expression in glioma.
Organizational Affiliation:
State Key Laboratory of Chemical Biology, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; Ben May Department for Cancer Research, The University of Chicago, Chicago, IL 60637, USA; Comprehensive Cancer Center, The University of Chicago, Chicago, IL 60637, USA.