The Discovery of MORF-627, a Highly Selective Conformationally-Biased Zwitterionic Integrin alpha v beta 6 Inhibitor for Fibrosis.
Harrison, B.A., Dowling, J.E., Bursavich, M.G., Troast, D.M., Chong, K.M., Hahn, K.N., Zhong, C., Mulvihill, K.M., Nguyen, H., Monroy, M.F., Qiao, Q., Sosa, B., Mostafavi, S., Smukste, I., Lee, D., Cappellucci, L., Konopka, E.H., Nowakowski, P., Stawski, L., Senices, M., Nguyen, M.H., Kapoor, P.S., Luus, L., Sullivan, A., Bortolato, A., Svensson, M., Hickey, E.R., Konze, K.D., Day, T., Kim, B., Negri, A., Gerasyuto, A.I., Moy, T.I., Lu, M., Ray, A.S., Wang, L., Cui, D., Lin, F.Y., Lippa, B., Rogers, B.N.(2024) J Med Chem 67: 18656-18681
- PubMed: 39446989
- DOI: https://doi.org/10.1021/acs.jmedchem.4c01851
- Primary Citation of Related Structures:
9CZ7, 9CZA, 9CZD, 9CZF - PubMed Abstract:
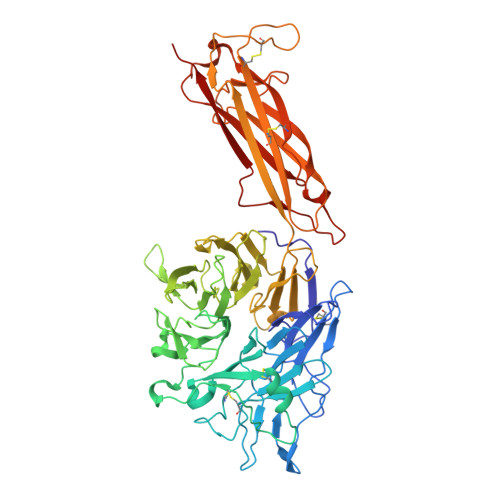
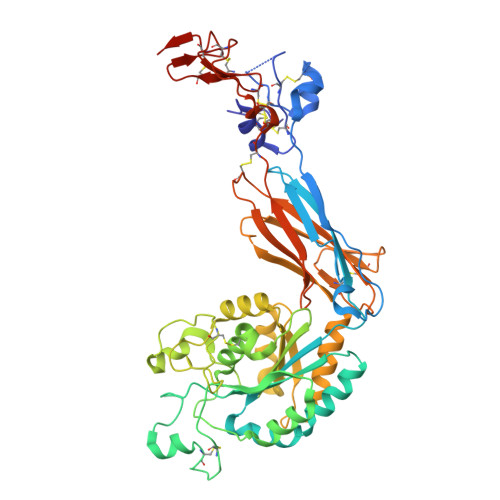
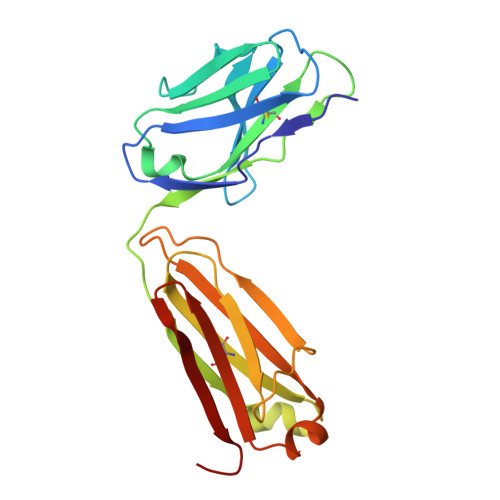
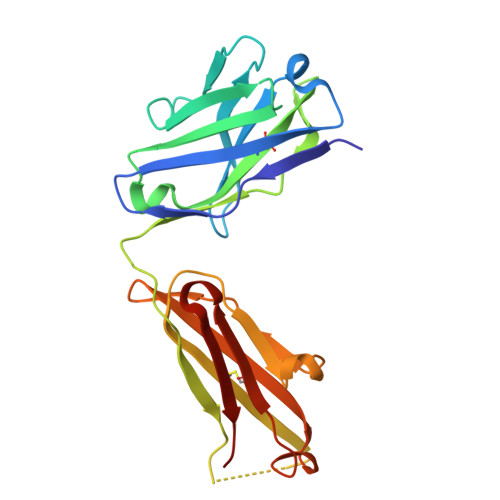
Inhibition of integrin αvβ6 is a promising approach to the treatment of fibrotic disease such as idiopathic pulmonary fibrosis. Screening a small library combining head groups that stabilize the bent-closed conformation of integrin αIIbβ3 with αv integrin binding motifs resulted in the identification of hit compounds that bind the bent-closed conformation of αvβ6. Crystal structures of these compounds bound to αvβ6 and related integrins revealed opportunities to increase potency and selectivity, and these efforts were accelerated using accurate free energy perturbation (FEP+) calculations. Optimization of PK parameters including permeability, bioavailability, clearance, and half-life resulted in the discovery of development candidate MORF-627, a highly selective inhibitor of αvβ6 that stabilizes the bent-closed conformation and has good oral PK. Unfortunately, the compound showed toxicity in a 28-day NHP safety study, precluding further development. Nevertheless, MORF-627 is a useful tool compound for studying the biology of integrin αvβ6.
- Chemistry, Morphic Therapeutic, Waltham, Massachusetts 02451, United States.
Organizational Affiliation: