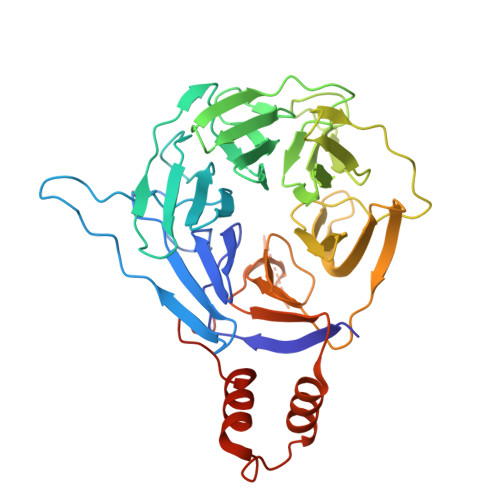
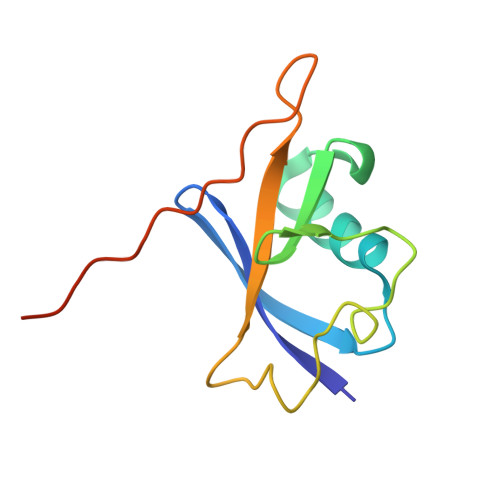
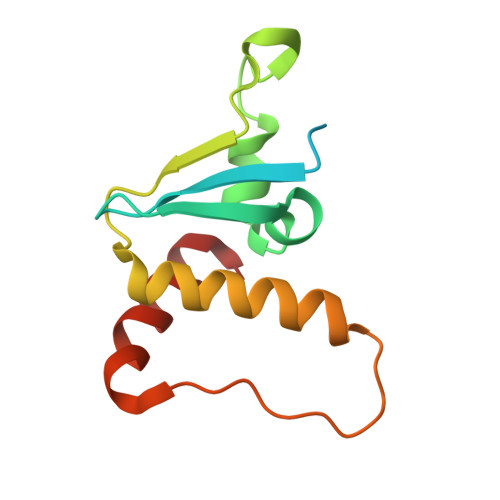
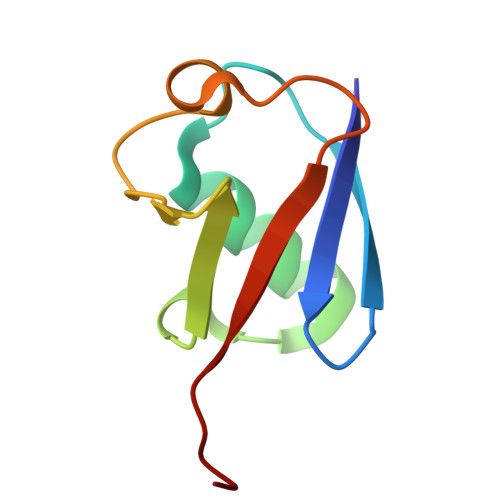
Structural basis for C-degron selectivity across KLHDCX-family E3 ubiquitin ligases
Scott, D.C., Chittori, S., Purser, N., King, M.T., Maiwald, S., Chuiron, K., Nourse, A., Lee, C., Paulo, J.A., Miller, D.J., Elledge, S.J., Haper, J.W., Kleiger, G., Schulman, B.A.To be published.